The virus battle
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 19 August 2022 | Inês Santos | No comments yet
Inês Santos explains how one can improve target quantification in testing for norovirus and hepatitis A with digital PCR.

Foodborne diseases are an increasing public health concern, resulting in a considerable number of deaths and hospitalisations, and significant economic losses due to food waste. The World Health Organization estimates pathogen-related foodborne illness affects 600 million people and causes 420,000 deaths worldwide every year.1
Viruses are one of the leading agents of food and waterborne outbreaks, representing 24.7 percent of all outbreaks in Europe in 2019. Of these, the enteric norovirus (NGI and NGII) is responsible for more widely reported and significant outbreaks (457 outbreaks with over 100 cases on average), although hepatitis A (HAV) virus, while less frequently transmitted, carries a high hospitalisation rate (73.3 percent in 2019).2 A lower number of reported outbreaks of enteric viruses was observed during 2020 and the first half of 2021, most likely due to a pandemic-driven reduction in surveillance indicators as well as the measures in place to combat COVID-19. However, recent data from the US Centers for Disease Control and Prevention (CDC) and the UK Health Security Agency reports a return to previously observed levels for these viruses.3-5
Both viruses can be transmitted through the faecal-oral route, but outbreaks are most commonly associated with the consumption of contaminated food. As these viruses can survive for several days at 4°C and up to several months in frozen products, the main sources of outbreaks from contaminated foods are related to the consumption of raw or barely cooked food, such as fruits, salads and vegetables, bivalve shellfish and water.
In foodstuffs, contamination is often linked with inappropriate handling by infected food workers or contaminated irrigation water; in shellfish, it is also associated with the bioaccumulation of viruses present in the water. Additionally, enteric viral agents are easily spread via fomites such as doorknobs and working surfaces, and individual devices such as phones or computer keyboards.6
Detection and quantification
The gold-standard protocol recommended for the detection and quantification of these pathogens is described in ISO 15216,7,8 comprising pre-processing according to sample type and an RT-PCR for each of the targets. The pre-processing is complex and time-consuming and yields small amounts of viral RNA, with an acceptable extraction efficiency level as low as one percent. Moreover, both the food matrices and the reagents used during sample processing lead to high carry-over of inhibitors into the amplification reaction.
Therefore, an adaptation of this procedure to a more sensitive and less prone to inhibition technology is desirable.
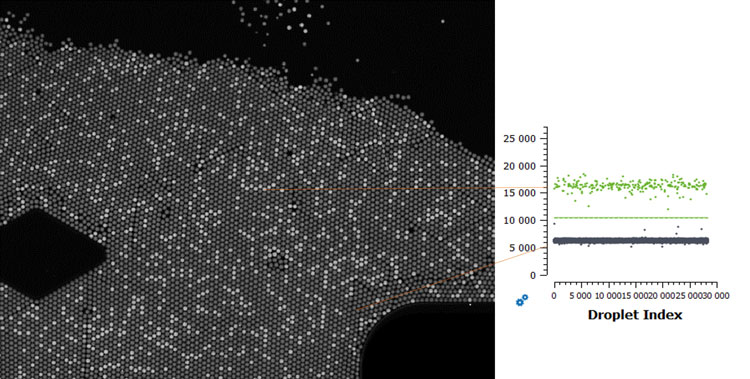

Figure 1: Image of a droplet digital PCR chamber for one target. Each droplet corresponds to one partition wherein PCR occurs. Brighter droplets correspond to positive partitions, where the target was detected, shown in green on the right 1-D plot
Digital PCR is a highly sensitive technology in which the sample is partitioned into thousands of small compartments wherein the amplification reaction occurs. Each compartment contains all the necessary reagents for the PCR, which follows the same principle of a real-time PCR. The results of each partition are then read individually, and a Poisson distribution is applied to obtain a quantification of the target detected against the number of partitions observed (Figure 1). This leads to a more robust method that is sensitive to low concentrations of the target and resilient to high levels of inhibitors, since they’ll be distributed among the partitions, which makes it particularly suitable for analysing food samples.
Contamination is often linked with inappropriate handling by infected food workers or contaminated irrigation water
Moreover, the partitioning facilitates the possibility of multiplexing, allowing the analysis of all four targets to occur within the same reaction, which saves time and the cost of reagents. Particularly for samples where the target abundance is low, a higher volume can be loaded into the reaction without inhibiting it, increasing the probability of detection. Lastly, and perhaps most importantly, the quantification obtained is an absolute quantification and does not rely on a calibration curve, which often skews the results.9
Here, we present a droplet digital PCR method developed by the SGS Competence Centre for Molecular Biology (CCMB) using Stilla® technologies. The method was created for the detection and absolute quantification of norovirus group I (NGI) and group II (NGII) and hepatitis A virus (HAV), along with mengovirus as an extraction control, in a single tetraplex reaction. It was developed according to ISO 15216, using the same target sequences and the same controls. It underwent optimisation and extensive validation according to the ISO guidelines submitted for digital PCR methods9 to ensure its fitness for purpose. Its development was performed according to the recommendations of the standard method in use, with every improvement made in comparison to the gold standard to ensure better results.
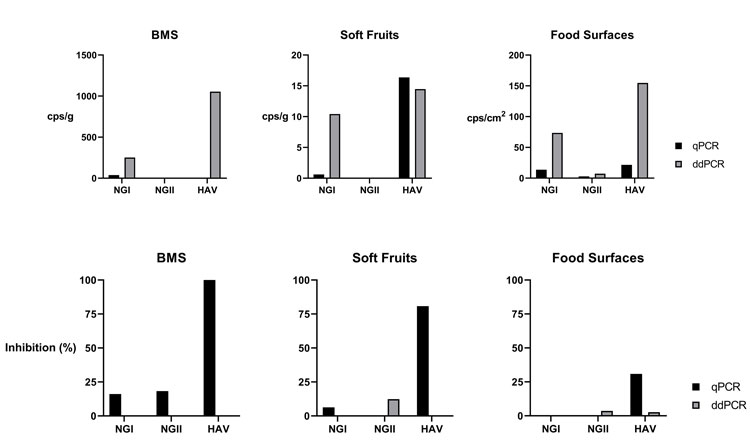

Figure 2: Comparison of results obtained for different matrices spiked with NGI, NGII and HAV, with the standard method (qPCR) and digital PCR (ddPCR). The top row refers to the quantification for each target per amount of sample obtained; in the lower row it is possible to observe the percentage of inhibition calculated
The method’s performance was evaluated following the established criteria, presenting an inclusivity and exclusivity of 100 percent, and a false negative rate and false positive rate of zero percent. The limit of detection (LOD), assessed with and without background RNA, was defined above three viral genome copies per microlitre of extracted RNA and limit of quantification (LOQ) above six viral genome copies per microlitre. The method was proven to be robust using a multifactorial approach. Statistical parameters for digital PCR quantification methods were applied as recommended, with the assessment of linearity, trueness and uncertainty meeting the acceptable criteria.9
You may also like:
PCR technologies for the detection of pathogens in the food industry
The method’s applicability in samples was tested by spiking the most commonly affected matrices – bivalve molluscan shellfish (BMS), soft fruits and food surfaces – with a mixture of certified reference material for NGI, NGII and HAV. RNA extraction was performed following a matrix-appropriate protocol, adapted from ISO 15216. Following the standard method’s recommendations, the extraction efficiency and possibility of inhibition were assessed as quality controls. All tests were performed in parallel using our digital PCR method developed in-house, and the standard real-time PCR method (Figure 2).
The results
As expected, target quantification was substantially more affected by the presence of inhibitors in real-time PCR, which in some cases resulted in the need to use diluted extracts, further lowering the method’s sensitivity. Target quantification was 84 percent higher overall for digital PCR than with the standard method, highlighting the more sensitive approach and resistance to inhibitors.
This method is the next to be released in a newly launched line of digital PCR kits for food safety and authenticity, developed by the Production Unit of SGS Molecular, along with CCMB.
About the author
Inês Santos is a researcher in the SGS Competence Center for Molecular Biology (CCMB). She has a Master’s degree in microbiology and has been working on molecular projects for several years. At the SGS CCMB she is responsible for the development and validation of various projects as well as for the implementation of different techniques.
References
- World Health Organization. 2015. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. World Health Organization.
- European Food Safety Authority and European Centre for Disease Prevention and Control. 2021. The European Union one health 2019 zoonoses report. EFSA Journal, 19(2), p.e06406.
- European Food Safety Authority and European Centre for Disease Prevention and Control. 2021. The European Union one health 2020 zoonoses report. EFSA Journal, 19(12), p.e06971.
- Centers for Disease Control and Prevention, CDC. (2022). NoroSTAT Data. Available at: https://www.cdc.gov/Norovirus/reporting/norostat/data. (Accessed: 20th June 2022).
- UK Health Security Agency. 2022. Routine Norovirus and rotavirus surveillance in England, 2021 to 2022 season. National Norovirus and rotavirus bulletin.
- Richards GP. 2016. Shellfish-associated enteric virus illness: Virus localization, disease outbreaks and prevention. In Viruses in foods (pp. 185-207). Springer, Cham.
- ISO 15216-1:2017 Microbiology of the food chain — Horizontal method for determination of Hepatitis A virus and Norovirus using real-time RT-PCR – Part 1: Method for quantification.
- ISO 15216-2:2019 Microbiology of the food chain — Horizontal method for determination of Hepatitis A virus and Norovirus using real-time RT-PCR – Part 2: Method for detection.
- ISO 20395:2019(E): Biotechnology — Requirements for evaluating the performance of quantification methods for nucleic acid target sequences — qPCR and dPCR.









