Science of Sake
Posted: 10 October 2016 | Caitlin E. Devor | No comments yet
This article outlines recent research on the brewing yeast of Japanese rice wine and highlights the unknown genetics of food microbes.


The basics of sake
Sake is made from five basic ingredients: rice, water, fungus, yeast, and time. Researchers recently identified a genetic vulnerability in the yeast responsible for producing some of the world’s highest-quality sake.1 The results highlight the vast potential to increase the scientific and food communities’ understanding of the yeast responsible for economically important foods and drinks.
Two microbes are essential for the chemical reactions that produce sake. One reaction is performed by enzymes produced by filamentous fungi, generally a species of Aspergillus, which converts rice starch into glucose. Budding yeast, generally a strain of Saccharomyces cerevisiae, perform another reaction to turn the glucose from rice starch into ethanol and carbon dioxide. The reactions performed by fungi and yeast occur simultaneously during the main fermentation period, a process called parallel fermentation. The speed and the by-products of the yeast’s chemical reactions create the final flavour of the sake.
The scientific community has exhaustively documented the yeast used in laboratory experiments to study cell development or genetic inheritance. Research yeast often has two sets of chromosomes, similar to humans, but food yeast can have three or four copies of each chromosome. The biochemical signalling pathways of research yeast usually produce minimal by-products, but food yeast are valuable because of their abundant and diverse by-products. Specific attributes of individual food yeast may be known, but an advanced understanding of their susceptibility to disease or deformity has not yet been documented.


A traditional sugidama, cedar ball, hangs at the entrance to the National Research Institute of Brewing in Saijo, Hiroshima, Japan. Cedar trees were felled to build the wooden brewing tanks originally used to ferment sake. Needles from the cut trees were arranged into a ball and the colour of the needles, changing from green to brown, indicated how long the sake had been brewing. Modern breweries use metal brewing tanks, but sugidama remain a symbol of sake at breweries, pubs, and liquor stores across Japan. Image by National Research Institute of Brewing
The current studies on sake yeast grew from the researchers’ primary interest in cell division. During the process of dividing from one cell into two, cells pause to check and repair any genetic irregularities caused during DNA replication or abnormalities of the mitotic spindle. The mitotic spindle is part of the cell’s internal architecture and moves chromosomes around the cell as it divides, like a microscopic version of the theatrical rigging that lets actors fly across a stage. The spindle assembly checkpoint ensures that both new cells receive the correct number of chromosomes.
“‘Checkpoint’ is a key word in both basic and applied science. Defects in a cell’s ability to correctly complete these checkpoints can be a cause of cancer in humans or ruined final food product in yeast,” says Dr Dai Hirata, a Visiting Professor at Hiroshima University and the Director/Department Manager of Asahi Sake Brewing Company in Niigata, Japan. Hirata is a molecular biologist, last author of two recent studies1,2 on sake yeast, and a trained sake taster with experience serving as an official judge at sake evaluation events.
The quality of sake
Two major factors that determine sake quality are: 1) the polishing ratio of the rice used to make the sake; an indicator of how much of the original brown rice has been removed to produce the polished white rice, and 2) the yeast chosen for the fermentation. Unsaturated fatty acids and proteins in the rice grains’ outer layers reduce sake quality, so removing these layers creates better taste. Rice polishing is time consuming and labour intensive, which is reflected in the higher price of sake made from highly polished rice. Different brewing yeast, whether for beer, wine, or sake, create different tastes in the final product due to factors such as how they make the sugar-to-alcohol conversion and the by-products that they release as part of many biosynthetic pathways. Sake yeast that produces fruity or floral scents and flavours is considered higher quality. The Japanese National Tax Agency maintains official standards for sake quality labelling.
Very high-quality types of sake are called daiginjo-shu and junmai-daiginjo-shu – made from rice with over half of the outer layers polished off – and are brewed for a long fermentation period at a low temperature before being filtered and bottled. The yeast strain Kyokai Number 1801 (K1801) has been used widely to make these high-quality sake. The major flavour components produced by K1801 are ethyl caproate, which has an apple flavour, and isoamyl acetate, which has a banana flavour. K1801 produces a low amount of total acids as it brews, which creates the smooth taste of its sake. Additionally, K1801 does not produce a foamy layer while brewing, meaning it requires less physical labour for brewers during the cleaning process between batches. With some yeast strains foam develops as yeast cells cling to carbonation bubbles during fermentation. Sake yeast designated with -01 lack a specific protein on the surface of their cells, which prevents them from clinging to the bubbles, meaning the carbon dioxide bubbles pop quickly and do not develop into a foam3.
Japanese sake brewers usually purchase yeast from suppliers annually at the start of the winter brewing season to ensure consistency in their product from year-to-year. The Brewing Society of Japan (BSJ) holds the rights to K1801 and sends the yeast to licensed brewers in slants, standard microbial transport test tubes. Usually, companies based overseas are not able to purchase yeast from the BSJ.
Sake yeast biology
A team of biologists based at Hiroshima University and the National Research Institute of Brewing (NRIB) in the historic sake brewing town of Saijo, identified a genetic mutation in K1801 that could ruin the brew of these high-quality sake. The research involved an academia-government-industry collaboration that has now produced two scientific publications.1,2
A previously identified mutation (FAS2-G1250S) in K1801 is a desirable change that makes the yeast produce high amounts of ethyl caproate. The research team recently identified a different mutation (CDC55-R48P) in a gene responsible for a spindle assembly checkpoint.1 This mutation poses a
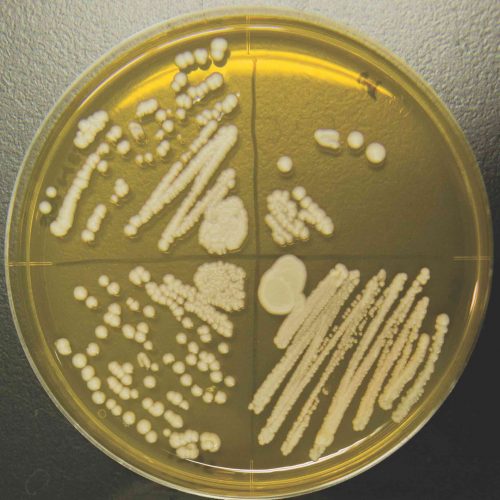

Colonies of the K1801 sake yeast growing in the laboratory at Hiroshima University.
potential risk for brewers because it can lead to instability of the yeast’s fermentation properties. If the spindle assembly checkpoint fails, the K1801 cells could develop genetic abnormalities that negatively affect their brewing qualities.
“Instability of K1801 on an industrial scale has not been detected at any time during the past decade since brewers began using the yeast. Indeed, K1801 has been used widely. However, in the future investigation of risk-free K1801 with normal checkpoint integrity will be important for high-quality sake brewing,” said Dr Naoto Okazaki of the BSJ.
The research on K1801 was made possible by an ongoing, not-yet-published project by Dr Takeshi Akao of the NRIB to sequence the full genomes of 23 species of sake yeast. Compiling such a database provides a resource for applied food researchers and basic biologists to explore the science of food yeast by identifying potential hazards and opportunities to enhance or repurpose the microbes.
In the latest publication by the research team, Dr Tetsuya Goshima of the NRIB and collaborators verified that K1801’s two mutations are not functionally related; a risk-free strain of K1801 should still produce a high level of delicious flavour components.1 The experiments included genetic tests, chemical analysis, and computer-assisted microscopic visual inspection of the yeast cells using a software programme designed to scan the physical shape of any cell sample and quantify the mean and the variance of the cells’ morphology, or physical appearance. This CalMorph software programme was developed by Dr Yoshikazu Ohya, Professor at the University of Tokyo, and other collaborators.4


Industrial-scale
sake brewing facility at the National Research Institute of Brewing. Brewers monitor fermentation primarily by measuring specific gravity and alcohol levels. Traditional breweries make sake in winter when rice is available from the autumn harvest and when cool air maintains the temperature of the facility.
Goshima and collaborators built a genetically engineered version of K1801 that had normal growth but maintained high production of ethyl caproate and used it to create a small-scale test brew of sake using 100 grams of rice in the NRIB laboratory.
“Our small-scale brew indicated that this version of the yeast without the growth-related mutation should maintain the high quality expected of daiginjo-shu,” said Hirata.
Future of sake yeast
Researchers successfully built a single-mutant version of K1801 without the spindle assembly checkpoint defect by inserting a non-mutant version of the gene taken from another standard sake yeast strain. However, brewers will continue to use the double-mutant K1801 because the Japanese market will not accept sake made from genetically modified yeast. The next step for the research team is to screen potentially thousands of K1801 yeast cells until they find a naturally occurring single-mutant with only the desirable high ethyl caproate mutation. Goshima is currently creating a positive-screening method to efficiently identify such a yeast at the NRIB.
Researchers anticipate that they have a high likelihood of success to find such a spontaneous mutant because of promising results in another strain of yeast called Ginjyo Number 9 (G9).2 Standard G9 has neither the spindle assembly checkpoint nor high ethyl caproate mutations, but Hiroyasu Tamura of the Asahi Sake Brewing Company and collaborators bred yeast under specific conditions to encourage the survival of certain natural mutants.2 They isolated a spontaneous FAS2-G1250S single mutant, designated G9CR, which showed high ethyl caproate-producing ability and maintained an intact spindle assembly checkpoint mechanism.
In an industrial-scale 600-kilogram test brew, they confirmed that G9CR does produce higher-quality sake than G9. Isolation of any spontaneous mutant with normal spindle assembly checkpoint integrity is an important verification of the validity of using screening methods like these to breed risk-free industrial yeasts. However, G9CR cannot equal K1801’s other excellent brewing properties, so the search for a K1801 single mutant will continue.
Identifying a naturally occurring single-mutant version of K1801 will ensure the reliable future of daiginjo-shu and junmai-daiginjo-shu for sake admirers around the world. Researchers involved in this project are located at Hiroshima University, the National Research Institute of Brewing (Japan), the Asahi Sake Brewing Company (Niigata), the Brewing Society of Japan, The University of Tokyo, The University of Pennsylvania, and Iwate University.
Find more Hiroshima University news on our Facebook page:
www.facebook.com/HiroshimaUniversityResearch
Asahi Sake Company in Niigata:
National Research Institute of Brewing:
About the author
Caitlin E. Devor is a Science Communicator originally from Pittsburgh, Pennsylvania, USA and a former member of Australia’s Science Circus where she regularly held fire in her hands, safely with science. She studied biology at Allegheny College and earned a M.S. in Science Communication from Australian National University. She is currently based in Hiroshima, Japan and writes to excite non-expert audiences about the daily-life relevance of new scientific accomplishments.
References
- Goshima T, Nakamura R, Kume K, Okada H, Ichikawa E, Tamura H, Hasuda H, Inahashi M, Okazaki N, Akao T, Shimoi H, Mizunuma M, Ohya M, Hirata D. Identification of a mutation causing a defective spindle assembly checkpoint in high ethyl caproate-producing sake yeast strain K1801. Bioscience, Biotechnology, and Biochemistry (2016) 80:8, 1657-1662. DOI: 10.1080/09168451.2016.1184963
- Tamura H, Okada H, Kume K, Koyano T, Goshima T, Nakamura R, Akao T, Shimoi H, Mizunuma M, Ohya Y, Hirata D. Isolation of a spontaneous ceruleninresistant sake yeast with both high ethyl caproate-producing ability and normal checkpoint integrity. Bioscience, Biotechnology, and Biochemistry (2015) 79:7, 1191-1199. DOI: 10.1080/09168451.2015.1020756
- Miyashita K, Sakamoto K, Kitagaki H, Iwashita K, Ito K, Shimoi H. Cloning and analysis of the AWA1 gene of a nonfoaming mutant of a sake yeast. J Biosci Bioeng. (2004) 97:1, 14-18. DOI: 10.1016/S1389-1723(04)70158-9
- Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, Suzuki G, Watanabe M, Hirata A, Ohtani M, Sawai H, Fraysse N, Latgé JP, François JM, Aebi M, Tanaka S, Muramatsu S, Araki H, Sonoike K, Nogami S, and Morishita S. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. (2005) 102, 19015-19020. DOI: 10.1073/pnas.0509436102
Issue
Related topics
Beverages, Genetic modification (GMO), Ingredients, Processing, Research & development
Related organisations
Asahi Sake Brewing Company, Brewing Society of Japan, Hiroshima University, Iwate University, The National Research Institute of Brewing, The University of Tokyo, University of Pennsylvania









