Salmonella serotyping – from reference to routine lab
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 3 May 2021 | Andrew Tomlin | No comments yet
Historically a mainstay of the reference laboratory, is Salmonella serotyping now possible in the routine micro setting?

Salmonella causes millions of human infections across the globe each year, with an estimated annual cost in the billions of pounds.1 In order to tackle this issue, regulations such as the EU legislation EC201/2003 have highlighted the control of Salmonella throughout the food production process as a critical issue, from production to consumption.
Through the use of good manufacturing practices and other food safety initiatives such as the British Lion Scheme, instances of Salmonella infection can be reduced and the impact limited. Considering the ever-growing increase in food production, the rise of antimicrobial-resistant Salmonella and the severity of typhoidal strains (the causative agent of enteric fever), the surveillance, control and tracking of Salmonella outbreaks is of crucial importance to public health.
What are Salmonella spp.?
Usually, Salmonella spp. reside in the intestinal tract of warm-blooded mammals, which are then shed through faeces or during slaughter. A variety of foods have been identified as vehicles for transmission, including fruit, vegetables, chocolate and milk. The human infection caused by Salmonella spp. is referred to as Salmonellosis, with non-typhoid Salmonellosis accounting for the majority of human infections. The infection usually lasts between four and seven days, includes the usual array of uncomfortable food poisoning symptoms, and is self-limiting in most cases.
There is a good degree of seasonality to Salmonella infections, with the summer months seeing the highest numbers in the UK.2 This is due to an array of factors, including higher ambient temperatures and seasonal consumer habits (such as barbecues and holiday travel) and is something that we also see reflected in detections in the laboratory setting. Another seasonal variation is the rise in Salmonella detections during the festive period, as the production and consumption of turkey spikes. While Salmonella do not form spores, they can survive in food for long periods and as one of the primary causes of foodborne illness, the detection, or verified absence in food, is vitally important for food business operators (FBOs).
Testing options
A genus of bacteria from the Enterobacteriaceae family, Salmonella spp. are gram negative, flagellated, rod-shaped bacteria that possess characteristic O and H antigens. Salmonella detection is often carried out using the reference method (ISO6579-1, 2017), or an alternative method validated against this, such as ELISA or PCR-based systems. Presumptive results obtained from an initial screening test are then confirmed as Salmonella species using biochemical tests such as an API20E, or a high-throughput system such as a MALDI Biotyper, both of which provide a ‘Salmonella sp.’ result. From a food testing point of view this type of result is usually sufficient, as absence of Salmonella is mandated by food safety regulation in most cases.
While this level of resolution is sufficient from a regulatory perspective, these tests do not have the ability to discriminate between Salmonella spp., which can be of paramount importance when carrying out source-tracking investigations, establishing potential links between Salmonella detections and in the poultry industry where absence of specific strains must be demonstrated. To acquire this level of information, further discriminatory analysis must be performed.
Challenges of Salmonella
The genus Salmonella comprises two species: Salmonella bongori and Salmonella enterica. While both species have the capacity to cause human illness, Salmonella bongori is mostly associated with cold-blooded animals and so the majority of emphasis in scientific literature and study is placed on S. enterica.
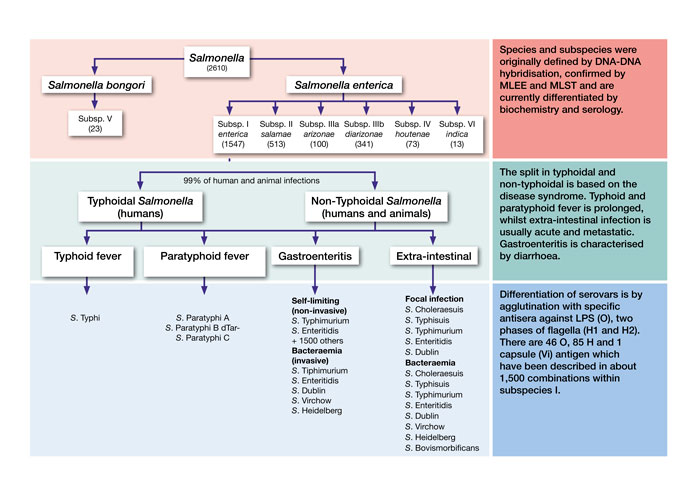

Figure 1: Taxonomy of Salmonella, adapted from Achtman, et al, 20124
Beyond species level, the taxonomy of S. enterica is fairly complicated due to the extensive array of serotypes, of which there are more than 2,600. A serotype is defined as a ‘serologically distinguishable strain of a microorganism’, or more simplistically, a group of microorganisms within a single species. Figure 1 displays the taxonomy of Salmonella. Of the six main S. enterica subspecies, Salmonella enterica subsp. enterica (I) is the largest and is responsible for approximately 99 percent of human infection.3
Salmonella serotyping
Traditionally, Salmonella isolates are separated into serotypes based on structural differences on the surface of the cells (O antigens) and thread‑like portions of the flagella (H antigens), using the Kauffman-White classification scheme. In this technique, antibodies are prepared against these specific antigens in a blood serum known as antiserum. Confirmed Salmonella sp. isolates are then tested with this antisera and are observed for agglutination reactions.
Through testing unknown samples against a series of antisera, the specific serotype of an isolate can be discerned. As previously discussed, there are a great number of serological variants of Salmonella and so this process can be very long and labour intensive, requiring highly experienced staff with a vast library of antisera at their disposal. Because of this, the Kauffman-White serotyping method is often only carried out by reference laboratories, with routine microbiology laboratories only stocking a small number of antisera.
As an example, at ALS Rotherham we stock the antisera for our in-house control strain, Salmonella enterica subsp. enterica ser. Nottingham, which enables us to distinguish our strain from others using the antisera O16, Henz15 and Hd. This serotype is recommended by the health protection agency in the UK for use as a control strain, due to being a very rare serotype and thus very unlikely to be isolated as a wild type. Historically, when further analysis was required for one of our samples, the isolates would be subcontracted to a reference laboratory capable of full serological testing. For a plethora of reasons, this type of analysis all too often had a lengthy turnaround time which, while accurate, was often too little too late and unhelpful in making a practical difference to our client, the FBO (food business operator).
New developments
In response to the status quo, several different methodologies have recently been developed that make serotyping more accessible and enable other organisations aside from reference labs to meaningfully serotype isolates.
One such system is the Check-Points Check & Trace (CTS) Salmonella assay, which has recently been implemented at ALS Rotherham. This system is a rapid genetic test based on a microarray (sandwich ELISA) platform, which can deliver results in as little as one day. Each position on the microarray represents a specific DNA marker associated with a unique Salmonella target sequence. Spots only become visible if the DNA markers exactly match the corresponding DNA sequences of the Salmonella isolate.
The combination of present and absent spots yields a specific pattern for each Salmonella serotype. The CTS assay confirms all Salmonella and identifies over 300 of the most common serotypes. Patterns not present in the validated database are assigned a genovar score, thereby allowing for direct comparison between isolates regardless of whether they’ve been assigned a serotype. Isolates not in the database can be sent to Check-Points for further analysis, which has often provided useful insight into the nature of the isolate.
A rapid serotyping method enables our business to swiftly serotype confirmed Salmonella isolates; as a group, ALS UK has been able to internalise previously subcontracted serotyping testing, which has reduced costs and turnaround times significantly. This has enabled clients to respond quickly and decisively to emerging Salmonella outbreaks, providing crucial information to assist them in making informed decisions to reduce and limit the impact to public health and their business.
About the author
Andrew Tomlin works as part of the Specialist Microbiology Team at ALS Laboratories, carrying out challenge tests, validations, method development and other bespoke microbiological and molecular testing projects, where he has worked for five years since obtaining his MSc in Biotechnology.
References
1. Oh J, Park M. 2017. Recent Trends in Salmonella Outbreaks and Emerging Technology for Biocontrol of Salmonella Using Phages in Foods: A Review. J. Microbiol. Biotechnol. , 27(12), pp. 2075-2088
2. Cherrie MPC, et al. 2018. Pathogen seasonality and links with weather in England and Wales: a big data time series analysis. BMC Public Health
3. Jajere SM. 2019. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. world, 12(4), pp. 504-521
4. Achtman M, et al. 2012. Multilocus Sequence Typing as a Replacement for Serotyping in Salmonella enterica. PLOS Pathog., 8(6).









