Appropriate controls for PCR detection of foodborne pathogens
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 30 April 2012 | Martin D’Agostino, Microbiologist, The Food and Environment Research Agency | No comments yet
Over the years, there has been a great increase in the number of PCR based assays for foodborne pathogen detection. For example, a very basic search for ‘salmonella food PCR assay’ using the PubMed.gov database will produce over 600 results. Clearly, this has led to a huge choice of PCR-based detection methods for analysts. This is especially so for analytical laboratories who choose to use non-proprietorial PCR-based methods, as opposed to commercially available complete PCR detection systems.
Since PCR-based assays are based on nucleic acid amplification, they are highly efficient, but they can also be negatively affected by the presence of food matrix-derived substances which can interfere or prevent the reaction from performing correctly. This is the case whether using a commercially available system, or a freely available non-proprietorial published method. Therefore, the use of appropriate and careful sample treatment must be applied or used to remove these inhibitory substances as much as is possible.
It must be noted however, that no sample treatment can be relied on completely, thus a suite of controls should be employed to verify that both the sample treatment and the PCR-based assay has performed correctly.
Over the years, there has been a great increase in the number of PCR based assays for foodborne pathogen detection. For example, a very basic search for ‘salmonella food PCR assay’ using the PubMed.gov database will produce over 600 results. Clearly, this has led to a huge choice of PCR-based detection methods for analysts. This is especially so for analytical laboratories who choose to use non-proprietorial PCR-based methods, as opposed to commercially available complete PCR detection systems. Since PCR-based assays are based on nucleic acid amplification, they are highly efficient, but they can also be negatively affected by the presence of food matrix-derived substances which can interfere or prevent the reaction from performing correctly. This is the case whether using a commercially available system, or a freely available non-proprietorial published method. Therefore, the use of appropriate and careful sample treatment must be applied or used to remove these inhibitory substances as much as is possible. It must be noted however, that no sample treatment can be relied on completely, thus a suite of controls should be employed to verify that both the sample treatment and the PCR-based assay has performed correctly.
Over the years, there has been a great increase in the number of PCR based assays for foodborne pathogen detection. For example, a very basic search for ‘salmonella food PCR assay’ using the PubMed.gov database will produce over 600 results. Clearly, this has led to a huge choice of PCR-based detection methods for analysts. This is especially so for analytical laboratories who choose to use non-proprietorial PCR-based methods, as opposed to commercially available complete PCR detection systems.
Since PCR-based assays are based on nucleic acid amplification, they are highly efficient, but they can also be negatively affected by the presence of food matrix-derived substances which can interfere or prevent the reaction from performing correctly. This is the case whether using a commercially available system, or a freely available non-proprietorial published method. Therefore, the use of appropriate and careful sample treatment must be applied or used to remove these inhibitory substances as much as is possible.
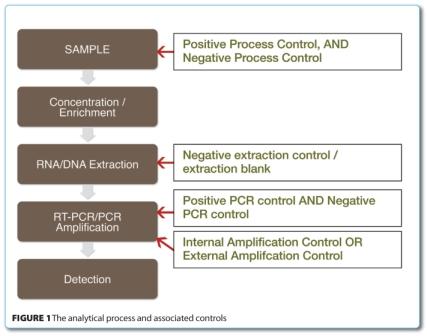
It must be noted however, that no sample treatment can be relied on completely, thus a suite of controls should be employed to verify that both the sample treatment and the PCR-based assay has performed correctly. The following controls which will be discussed here with their brief descriptions can be found in ISO 22174:2005 Microbiology of food and animal feeding stuffs – Polymerase chain reaction (PCR) for the detection of food-borne pathogens – General requirements and defini – tions section 3.5. Figure 1 outlines the process a sample goes through for the detection of foodborne pathogens (bacteria or virus), using PCR as an end-point detection, and the stages where the controls are required.

Other ISO standards are also available which should be considered:
- ISO 20837:2006 – Microbiology of food and animal feeding stuffs. Polymerase chain reaction (PCR) for the detection of foodborne pathogens. Requirements for sample detection for qualitative detection. Refers to ISO 22174 for controls
- ISO 20838:2006 – Microbiology of food and animal feeding stuffs. Polymerase chain reaction (PCR) for the detection of foodborne pathogens. Requirements for amplification and detection for qualitative methods. Refers to ISO 22174 for controls
- ISO 22118:2011 – Microbiology of food and animal feeding stuffs. Polymerase chain reaction (PCR) for the detection of foodborne pathogens. Performance charac – ter istics. Refers to ISO 22174 for controls. Includes requirements for validation.
- ISO 22119:2011 – Microbiology of food and animal feeding stuffs. Real-time polymerase chain reaction (PCR) for the detection of foodborne pathogens. General require – ments and definitions. Refers to ISO 22174 for controls; ISO 22119 specifically mentions the internal amplification control
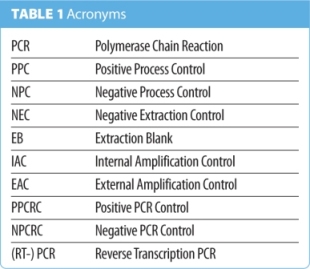
At this point, I would like to say a word about the use of acronyms. They can get confusing, and there are many within the field of molecular diagnostics, but they are unavoidable. Table 1 gives a summary of the acronyms used in this article.

Controls for the sample treatment step
To be able to control both sample treatment and detection, controls need to be included at the beginning of the actual sample treatment. These controls are the Positive Process Control (PPC) and the Negative Process Control (NPC).
The Positive Process Control (PPC) ensures that a sample is not declared target microorganism-free due to failure of the sample treatment. It is a sample, spiked with the target microorganism, which should be treated in the same way as the test samples. Its purpose is to verify that the sample treatment has functioned correctly, and identify those samples in which sample treatment has failed. Interpretation: If there is no signal obtained for the PPC, and all test samples are negative, it means that the sample treatment has failed.
The purpose of the Negative Process Control (NPC) is to verify that the sample treatment reagents and/or equipment are not contaminated with the target and/or its amplicon. It is a ‘sample’ which does not contain any matrix so, in place of the sample which would have been tested could be the same weight / volume of sterile distilled water. This control is processed exactly the same as a real test sample would be. An NPC must be used with every batch of sample analysed. Interpretation: There must be no target signal obtained from the NPC. If there is a signal, then contamination has occurred and we cannot trust the results of the actual test sample. In this case, the whole process should be repeated.
Control for the nucleic acid extraction step
Negative Extraction Control (NEC) / Extraction Blank (EB). This control is target free, so it is just water which is carried through all steps of the nucleic acid extraction procedure. It is used to ensure that the extraction process has not been contaminated with the target or its amplicon. Interpretation: If the target signal is present in the NEC / EB, it means the extraction reagents / equipment are contaminated. In this case, these reagents should be replaced and all equipment should be suitably decontaminated. It is not necessary to use this control if we are using an NPC.
Controls for the amplification step
It is equally important to be able to control the performance of the PCR assays, and so controls have to be included in the (RT-) PCR reagent mixtures, or additional reactions have to be performed. These controls include either an Internal or External Amplification Control (IAC or EAC) and a negative and positive amplification control (NAC and PAC).
Internal Amplification Control (IAC)
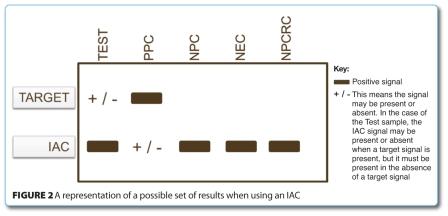
Perhaps one of the most important controls which should be incorporated into a food PCR based assay is an internal amplification control which is a non-target DNA / RNA sequence present in the very same reaction as the sample or target nucleic acid extract. If it is successfully amplified to produce a signal, any non-production of a target signal in the reaction is considered to signify that the sample did not contain the target pathogen or organism – a true negative. If however the reaction produces neither a signal from the target nor the IAC, it signifies that the reaction has failed. This may be due to inhibitory substances, or it may be due to reaction failure1,2. There may be several reasons why reaction failure has occurred apart from inhibition:
- Human error may have been the cause – was the sample really added?
- The thermocycler may not be functioning correctly3
- One of the PCR reagents may not have been added
- The sample may not have been loaded into the gel / well correctly depending on whether we are using conventional or real-time PCR
Whatever the reason may be, if a sample does not contain an IAC, there is no way of knowing whether a negative signal response is the cause of inhibition, a failed reaction, or if it is a true negative, and that may have serious consequences when analysing for potentially harmful foodborne pathogens. Figure 2 shows a representation of a possible set of results when using an IAC.

External Amplification Control (EAC)
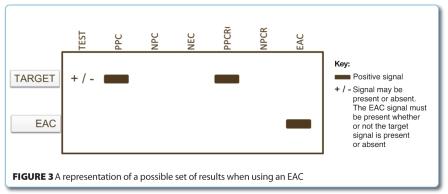
ISO 22174 includes the option of using an EAC. The EAC is used alternatively to an IAC. Its purpose is to verify reactions which have functioned correctly, and identify those which have been inhibited; in effect, the same function as the IAC. The incorporation of an external amplification control (EAC) aids in the identification of possible inhibited reactions.
The EAC is not added to the same reaction as the test sample, hence the term ‘external’, but is included as a separate reaction for each batch of samples tested in a PCR run. There is an assumption that the reaction has performed with a similar efficiency to the test sample, so if a negative signal is obtained (i.e. no signal at all is obtained), then it signifies that the nucleic acid extract must contain inhibitory substances, and therefore this is the reason the reaction has failed.
I would argue, however, that it is actually impossible to tell by the use of an EAC what has happened in the test sample, since it is a completely separate reaction. It is possible that human error can be a factor in the reaction failure, as discussed previously, so my conclusion is that if no signal is obtained whatsoever, it is impossible to unambiguously define what has caused the failure. If an IAC signal is obtained, at least we definitely know that the amplification reaction has performed correctly and it must be the sample treatment which has been the problem. When no signal is obtained as is possible in the case of an EAC, we do not know whether it is due to the sample treatment, human error or thermocycler failure. We cannot rely purely on a positive amplification control to give that answer either, which is the control discussed next. Figure 3 shows a representation of a possible set of results when using an IAC.

Whether you should use an IAC or EAC is a debated subject. There are arguments for both. Some are concerned that because the IAC is contained within the same reaction as the sample, and in most cases, it uses the same target primers, competition will occur and sensitivity will be compromised. This could happen if the IAC concentration has not been carefully optimised. This potential problem is avoided if you use an EAC, however as we have discussed, using an EAC does not control for all situations which could occur such as individual pipetting errors. An additional concern of using an EAC is that it increases the number of reactions required for each sample, significantly adding to costs.
Positive PCR Control (PPCRC)
This control is a nucleic acid amplification reaction, into which a known amount of nucleic acid from the target pathogen is included and its role is to verify amplification performance of the target sequence. When an IAC or EAC is used, it can fulfil the role of a positive amplification control; however, when using a reaction involving a probe for the target, as used in real-time PCR, the PPCRC verifies probe performance. If conventional PCR is being used, it need only be included if using an EAC and if so, should be included in every batch of sample in a cycler run. Interpretation: A target signal must be obtained from the PPCRC.
Negative PCR Control (NPCRC)
The final control to be considered is the NPCRC. It is an amplification reaction in which water takes the place of the nucleic acid extract. It must include either an IAC, or an EAC as a control. The NPCRC verifies that the (RT-) PCR mastermix reagents are not contaminated with the target, or the PPC, or their amplicons. Interpretation: No target or PPC signal should be obtained from the NPCRC, however IAC / EAC signals must be obtained.
Discussion
There are several main controls required when performing a PCR-based method for detection of foodborne pathogens. It is important to realise that the results from each of these controls should not be considered in isolation. They should be considered in context with each other. For example, if the target signal is obtained but the positive process control signal is not obtained, we should not just assume that the result is acceptable. Also, consider the target signal may be a result of contamination, so we should also check the results of the Negative Process Control, the Negative Extraction Control / Extraction Blank, and the Negative PCR Control. If any of these show a target positive result, we should not accept the positive test result as it is possible it is due to contamination – a false positive.
An additional control which is not mentioned within ISO 22174 is termed a Sample Process Control (SPC). In this case, an SPC would be a non-target organism added to every test sample including to the negative control sample at the start of analysis, and must be detected in every sample into which it has been added. It should not be added to its associated ‘SPC-NC’ (Sample Process Control – Negative Control), and it requires a separate Internal Amplification Control to be constructed. The principle of an SPC is that if it is detected, then the method was performed correctly. If it is not detected, the method has failed and the foodstuff must be reanalysed. In addition to this qualitative interpretation of an analytical result, the SPC can also allow a determination of the recovery efficiency for each individual sample. This can be done by comparing the (RT-) PCR results of the SPC before and after addition to the sample. This control has already been used successfully in the detection of foodborne viruses4,5 and research is being done which utilises it in the detection of foodborne bacteria.
This suite of controls, should give the food analyst confidence that the results obtained from their PCR analyses are both accurate and unambiguous.
References
1. Hoorfar, J., et al (2003). Diagnostic PCR: Making internal amplification control mandatory. Journal of Clinical Microbiology 41 5835. [Journal of Applied Microbiology 96 221 – 222 (2004); Letters in Applied Microbiology 38 79 – 80. (2004)]
2. Hoorfar, J., et al (2004). Practical considerations in design of internal amplification control for diagnostic PCR assays. Journal of Clinical Microbiology 42 1863 – 1868
3. Anonymous ISO/TS 20836:2005 Microbiology of food and animal feeding stuffs — Polymerase chain reaction (PCR) for the detection of foodborne pathogens — Performance testing for thermal cyclers
4. D’Agostino, M., et al (2012) Multicenter Collaborative Trial Evaluation of a Method for Detection of Human Adenoviruses in Berry Fruit. Food Anal. Methods 5:1–7
5. D’Agostino, M., et al (2011) Nucleic acid amplification-based methods for detection of enteric viruses: definition of controls and interpretation of results’ Food Environ Virol 3:55–60
Issue
Related topics
Food Safety, Hygiene, PCR-based assays, Polymerase Chain Reaction (PCR)









