Mass spectrometry in food microbiology: identifying microbial isolates within minutes
Posted: 3 January 2019 | Erin Crowley | No comments yet
Time is often the most critical factor in food microbiology laboratories, so the ability of MALDI-TOF MS to quickly and accurately confirm and identify microbes is invaluable in routine testing, as Erin Crowley explains.

The identification and confirmation of microorganisms is a task that spans several industries, including pharmaceuticals, cosmetics, and food. With accurate microbiology testing at their disposal, manufacturers are able to make quick decisions with regard to quality and safety; for example to detect possible food spoilage organisms or foodborne pathogens, or monitor environmental pathogens or technological microflora. Regulations are constantly becoming more stringent in the food industry, with pressure applied not only from regulatory bodies, but customers as well, demanding rigorous testing on food products.
A range of organisms, including bacteria, yeasts, moulds and parasites, can contaminate food. Common foodborne pathogens include Salmonella spp., Cronobacter spp., Campylobacter spp., and Listeria monocytogenes, which are capable of causing serious disease outbreaks through contaminated food if not detected fast enough. Process hygiene and food quality are each assessed by monitoring quality indicators, such as Escherichia coli, Listeria spp, yeasts and moulds. A wide variety of technologies are used to identify these microbes in food, from traditional culture-based methods and molecular detection using polymerase chain reaction (PCR) assays, to increasingly sophisticated analytical technology such as mass spectrometry (MS). The ability to identify microorganisms at the genus or species level enables a deeper level of screening, and researchers and test manufacturers are always searching for more advanced methods for faster tests with higher accuracy and sensitivity.
The move to mass spectrometry
The food industry has become accustomed to AOAC- or ISO 16140-approved rapid testing methods. The qualitative/quantitative methods have been fully accepted by food safety authorities, and their adoption has also been acknowledged in regional regulations, encouraging their applications to the large varieties of food matrixes. The development of new technologies, such as matrix-assisted laser desorption/ionisation (MALDI) time-of-flight (TOF) MS or sequencing moves food microbiology testing to the 21st century, enabling faster and more reliable results.
Time is often the most critical factor in food microbiology laboratories, so the ability of MALDI-TOF MS to quickly and accurately confirm and identify microbes is invaluable in routine testing. Comparative studies have demonstrated the difference in percentage of correctly identified bacteria and fungi isolates using MALDI-TOF MS (100 per cent) and traditional identification panels (94-97 per cent), with speed and reagent costs also representing key benefits of MALDI-TOF MS.1
The Microbiology Food Laboratory at Q Laboratories (Cincinnati, Ohio) has recently adopted MALDI-TOF MS for its routine food microbiology testing and research and development (R&D) work. The Microbial Identification Laboratory uses a standard ‘tool box’ of methods, which includes standard benchtop culture methods, automated biochemistry, microscopic evaluation, and 16S rDNA sequencing. An internal review of the technology used for microorganism identification, and requests from clients for alternative methods, spurred the consideration of MS for identification work. At the time, the laboratory was outsourcing to specialist identification facilities, so the Technology Advancement Group (TAG) Team was formed, which is now specifically charged with reviewing those technologies currently in use and those to be implemented in the future.
The food microbiology work is split between routine testing – where clients submit samples for confirmation and identification of microorganisms in food products – and R&D – where clients wishing to market and sell a diagnostic assay require validation for certification.
Routine testing with MALDI-TOF MS
In January 2017, the Microbiology Food Laboratory added MALDI-TOF MS to its tool box for routine microbial identification and R&D. This technology determines a unique protein fingerprint of a microorganism, which is used to reliably identify a particular species by matching the fingerprint with thousands of reference spectra from a comprehensive library. This tool is complemented by existing techniques, such as sequence-based identification with rDNA sequencing methods.
This MS technology enables the laboratory to offer flexible options for turnaround time (TAT) and cost, to fit in with the client’s needs. This is particularly valuable as it creates a completely customisable service, and provides the option of same-day results for rapid pathogen identification, for example. This adaptability was not available with previous methods. The confidence in the results obtained with the MALDI-TOF system and increased sensitivity means that fewer re-tests are required, so when an identification is provided first time, the client can make decisions immediately.
Traditional biochemical tests would take 3-7 days in the case of pathogen contamination. In the case of presumptive positive detection of a pathogen, time is critical for the confirmation results, to avoid delay (i) in product recalls, potentially leading to an outbreak of infective microorganisms, or (ii) in non-confirming the pathogen presence, which then leads to food wastage from products with short shelf-lives. The Food Microbiology Laboratory now analyses samples with MALDI-TOF MS in 30 minutes to two hours, to either confirm presumptive results of an alternative method, or to identify an organism from a plate.
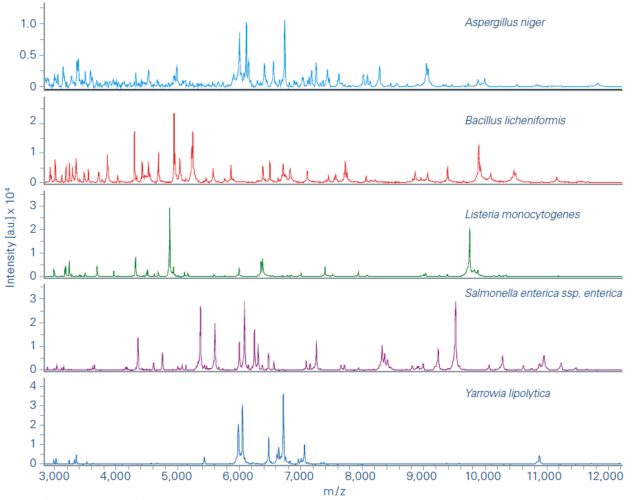

Figure 1: Example spectra obtained from the MALDI Biotyper (MBT) (Bruker Daltonics, Bremen).
Validating new analytical methods
While routine laboratory tests analyse samples from food products, the ‘samples’ that the R&D laboratory tests are the technologies themselves. Technology companies and diagnostic partners approach Q Laboratories with a new technology or assay that requires method validation – for example, for testing Salmonella, Listeria or Legionella – and request a proposal for the work. Q Laboratories establishes a study design with the client and the method validation organisation; ie, AOAC INTERNATIONAL in the US. The study should fulfil the AOAC Appendix J guidelines requirements.2
MicroVal and AFNOR Certification are the certification bodies in Europe, and the method performances are assessed using the ISO 16140 standard.3,4 The ‘Expert Laboratory’ (in this case, Q Laboratories) is the party that prepares the protocol, in collaboration with the technology provider, and submits the protocol to the certification body for the technical reviewers to approve. Q Laboratories is the only North American laboratory to be an Expert Laboratory for all three of the major certification bodies: AOAC, MicroVal and AFNOR Certification. This enables the design of combined AOAC/ISO validation studies and fulfilment of all client requirements.
Collaborative studies
In the case of an AOAC-OMA study or an ISO 16140 validation study, a comparison collaborative study takes place with one leading in-house laboratory, such as the Microbiology R&D Laboratory, challenging the reproducibility of a technology. The organising laboratory is responsible for recruiting 12-15 laboratories throughout the world, which have agreed to test the blind-coded samples sent to them. All laboratories evaluate the samples using the same technology, on the same day. Once the analysis is complete, the laboratories submit the data for the leading laboratory to conduct the statistical analysis and then prepare a journal-ready manuscript for the client and to submit to a certification body, where the technical committee review and approve by consensus. This approval means the method is fit for purpose and meets the validation requirements.
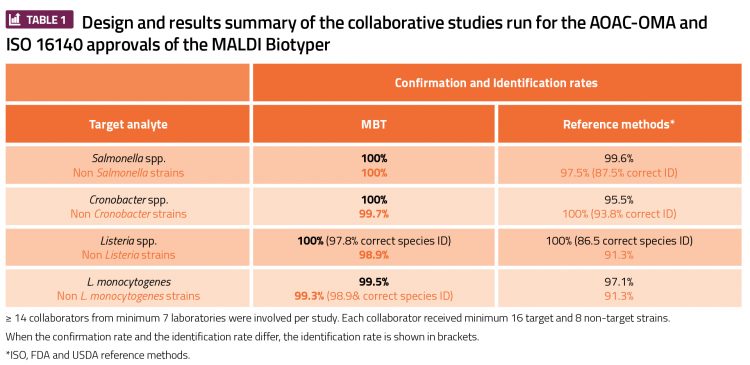
Q Laboratories conducted AOAC-OMA and ISO 16140 studies for the Bruker MALDI Biotyper (MBT). By combining existing guidelines, such as the AOAC Appendix J guidelines and the new ISO 16140 part 6 standard,4 an optimal study design was created to validate the use of the MBT for food microbiology analysis. These studies were the first of their kind, as the designs for validating a confirmation and identification method had not hitherto caught up with the newest technology.
Q Laboratories served as the lead laboratory for two separate studies, gaining AOAC-OMA status for the MBT in December 2017. The first was for confirmation and identification of Salmonella spp., Cronobacter spp. and other Gram-negative organisms (OMA#2017.09), and the second was for the confirmation and identification of Listeria monocytogenes, Listeria spp. and other Gram-positive organisms (OMA#2017.10) in food microbiology.5,6 Q Laboratories was also the expert laboratory for the ISO 16140 approvals by MicroVal. Pioneering studies were organised and four certificates were obtained in early 2018.7 Together with the MicroVal technical committee members, the proof of concept was presented with a poster and a symposium at the Internal Association of Food Protection (IAFP) event in Stockholm in April 2018.8,9
Having certification at the level of an AOAC Official Method and an ISO validated/MicroVal certified method is acceptable to the Food and Drug Administration (FDA) for pathogen confirmation and the European regulation EC 2073/2005.10 Both FDA and European regulations require a third-party certification body to have approved the validation data, and deem that as an equivalent alternative confirmation/identification method to their own methodology, which is considered the gold standard.

The future of microbial confirmation and identification
Mass spectrometry has historically been an analytical chemistry tool, and the reliability, robustness and reproducibility of the MBT in the hands of microbiologists has been recently demonstrated through the AOAC-OMA and ISO 16140 approvals. Integrating the MALDI-TOF MS into routine testing workflows results in a significant consolidation of resources, as it replaces multiple traditional and biochemical identification methods, and eliminates the burden of multiple steps, workstations and metrology requirements of DNA sequencing. There is no requirement to know the type of microorganism prior to analysis, as bacteria, yeast or mould samples are analysed together, following the same workflow.
Q Laboratories is expanding its confirmation and identification offering with MALDI-TOF MS and rDNA sequencing, and is looking to increase its environmental testing offering, as the need for identification of food and environmental contaminants continues to rise. From an isolated colony, the confirmation/identification result is obtained in 15 minutes for one isolate, and less than one hour is required for a full MALDI target, ie, 95 results. Having the technology onsite improves reactivity when time matters for decision-makers.
About the author
Erin Crowley is Chief Scientific Officer at Q Laboratories. She oversees the Microbiology Laboratory Services at Q Laboratories, and is a fellow of AOAC International. As Chief Scientific Officer, Erin has played an integral role in modernising the food microbiology laboratory, to streamline the workflow and make routine testing as efficient as possible.
References
- Elbehiry A, Marzouk E, Hamada M, Al-Dubaib M, Alyamani E, Moussa IM, AlRowaidhan A, Hemeg HA. (2017) Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiologica, Vol 40(4), pp 296-278.
- Official Methods of Analysis (2016) 20th Ed. AOAC INTERNATIONAL, Rockville, MD, Appendix J. http://www.eoma.aoac.org/app_j.pdf (accessed September 2017).
- ISO 16140-2:2016 ¾ Microbiology of the food chain – Method validation – Part 2: Protocol for the validation of alternative (proprietary) methods against a reference method.
- ISO/FDIS 16140-6:2017 ¾ Microbiology of the food chain – Method validation – Part 6: Protocol for the validation of alternative (proprietary) methods for microbiological confirmation and typing procedures.
- Bastin B, Bird P, Crowley E, Benzinger MJ, Agin J, Goins D, Sohier D, Timke M, Shi G, Kostrzewa M. 2018. Confirmation and Identification of Salmonella, Cronobacter spp., and Other Gram-Negative Organisms by the Bruker MALDI Biotyper Method: Collaborative Study. First Action 2017.09. Journal of AOAC International Vol. 101, No. 5, 2018.
- Bastin B, Bird P, Crowley E, Benzinger MJ, Agin J, Goins D, Sohier D, Timke M, Awad M, Kostrzewa M. 2018. Confirmation and Identification of Listeria monocytogenes, Listeria and Other Gram-Positive Organisms by the Bruker MALDI Biotyper Method: Collaborative Study, First Action 2017.10. Journal of AOAC International Vol. 101, No. 5.
- http://microval.org/en/issued-certificates/
- Bastin B, Bird P, Crowley E, Diep B, Ferro I, Hammack T, Jacobs H, Kostrzewa M, Le Doeuff C, Peron S, Rannou M, Sohier D, Timke M, in ’t Veld P, Witsenburg J. 2018. Validation of a confirmation method according to ISO/DIS 16140-6:2017 A MicroVal pilot study using the MALDI Biotyper as an alternative for Salmonella confirmation. International Association of Food Protection event, Stockholm, Sweden, April 2018. Poster P2-16.
- Arbault P, Sohier D, in ’t Veld P, Pryor J, Crowley E, et al. 2018. Turning Sequencing and Mass Spectrometry into Routine Testing Tools for Microbial Strain Characterization. International Association of Food Protection event, Stockholm, Sweden, April 2018. Symposium S19.
- COMMISSION REGULATION (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs.