Evaluation of ISO method 21527-1 compared with other yeast and mould detection systems
Posted: 28 May 2017 | Yiping Chen; Ruth Fowler | Institute of Life and Earth Sciences | EGIS | Heriot-Watt University | No comments yet
Yiping Chen and Ruth Fowler of the Institute of Life and Earth Sciences at Heriot-Watt University offer their insight.

Fungal contamination of food is a major problem, particularly in terms of food spoilage, but there is also the risk of spreading fungal pathogens to vulnerable people and contamination of agricultural food products by mycotoxin1-3. The Soleris system was initially developed in the 1990s to detect microbes in dairy products4,5 based on the growth of microbial contaminants in selective broths and the detection of pH changes induced by microbial metabolic by-products, causing colour changes in an agar indicator5. In this study, the automated Soleris 128 system was evaluated for its ability to detect yeast and moulds in various food matrices (one matrix of each food category defined by ISO). It detects carbon dioxide which is produced by fungal microbes growing in the system by measuring colour changes in an agar plug at the base of the vial. Only gases, such as carbon dioxide, can connect with the agar through a gas-permeable (and liquid-impermeable) membrane at the base of the vial6.
Materials and methods
The ability of the system was tested using four yeast species and two moulds in five different beverage and food samples plus one environmental sample. The challenge assays were: C. krusei in tap water (mimicking environmental sampling), S. cerevisiae in fruit juice, C. albicans in a tomato ketchup sauce, C. tropicalis in fruit yogurt, A. caseiellus mixed with deli ham and P. chrysogenum mixed with sliced smoked salmon. The food samples were purchased in a local supermarket and tap water collected from a local residence in Edinburgh city. All the solutions and media that were used in this study for plating were autoclaved at 121°C for 15mins. All the solutions and media were stored at 4°C.
Preparation of the fungi and bacteria
The microbes were inoculated into 9ml potato dextrose broth (Acumedia 7585A) and incubated at 25°C for at least two days. All the yeasts grew faster than the moulds; yeasts were ready to serially dilute within two days, whereas the moulds were sufficiently turbid after four days. Most of the bacteria used for the exclusivity testing (P. aeruginosa, P. fermentens, K. pneumoniae, E. coli and E. faecalis) were cultured overnight in 90ml Nutrient Broth (NB) (Oxoid CM0001) and L. brevis in 90ml of Mann Ragosa and Sharpe broth (MRS) (Oxoid CM0359) in a 250ml flask at 37°C in a shaking incubator.
Dilution and enumeration
To estimate the colony forming units per ml (cfu·ml-1) for the challenge experiments, a serial dilution in PBS (Acumedia 7380A) was prepared to 10-8 and 1ml from each dilution was spread on Dichloran Rose-Bengal Chloramphenicol (DRBC) agar (Acumedia 7591A) plates, incubated at 25°C for at least 48h. However, for the moulds, four or five days were needed to observe colonies on the agar. The same procedure was carried out for the bacteria used for the exclusivity assay, as described above, except that the incubation temperature was 37°C and nutrient agar (Oxoid CM0001) or M.R.S. agar (only L. brevis) (Oxoid CM0359) were used instead of DRBC agar.
Soleris system and software
At the beginning of each validation test, the appropriate verifications and settings for the test were carried out as per the manufacturer’s instructions. The results of the assays were recorded as times to a positive result and as traces of optical units over time. The positive results were recorded when the software registered three increases of 10 optical units, which corresponded to the threshold set at the creation of the test.
Spiking procedure
The spiking procedure was determined by the ISO 16140-2 method. Estimates of the volume required to achieve appropriate spike levels from diluted cultures were made from the results of the plate counts. In each Stomacher bag, 10g of food/ beverage sample (tomato sauce, fruit yogurt, fruit juice, deli ham, smoked salmon or tap water) and 90ml buffered peptone water (BPW) (Acumedia 7417A) were added and then the appropriate volume and dilution of the yeast or mould culture to achieve the different spiking levels (0cfu·ml-1 = unspiked sample, 1cfu·ml-1, 10cfu·ml-1) and mixed during 1 minute using a Stomacher. For the 0cfu ml-1 and 10cfu ml-1 five replicates were analysed. The 1cfu·ml-1 group was allocated at least 20 Soleris vials and 20 plates and fractionate results were expected. One exception to this rule was applied for sliced smoked salmon spiked with P. chrysogenum. This mould strain was difficult to inoculate accurately due to clumping. The experiment had to be repeated with a vortexing step. Therefore, two runs were conducted and the RLOD was calculated using data from both, so there were 10 replicates for 0cfu ml-1 and 10cfu ml-1 and 40 replicates for 1ml cfu-1. Antibacterial supplement (600μl) and 1000μl of the spiked sample was added to each vial. To each DRBC plate, 100μl of the spiked sample liquid was added, spread to form a lawn and then incubated at 25°C for five days. The vials were placed in the detection system according to the manufacturer’s instructions and as described above. The completed system results were available after 72h, although positive samples were indicated after three rises in CO2 levels at the preset threshold, usually within 48h, whereas the yeast and mould detection plates required an incubation of four days and five or more days respectively.
Exclusivity test procedure
To ensure that the detection method detected yeasts and moulds but not bacterial contaminants, 103-104cfu of P. aeruginosa, P. fermentens, K. pneumoniae, E. coli, E. faecalis and L. brevis were introduced into the Soleris yeast and mould system vials. As before, 600μl anti-bacterial supplement and 1000μl of the appropriate culture dilution was added to each vial, and assayed by the machine for 72h.
Results
This study aimed to test two aspects; to compare ability to detect fungi to the standard plating detection method (ISO 21527-1) to determine which method was more sensitive, and secondly to check that this system did not produce false positive results in the presence of bacteria.
Detection of yeasts and moulds
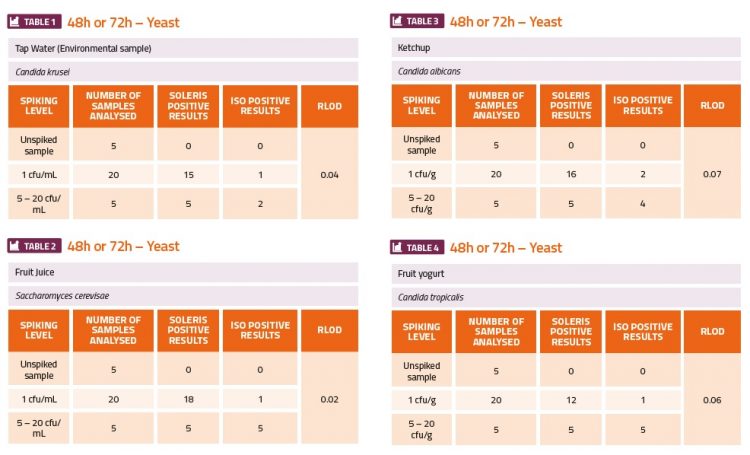
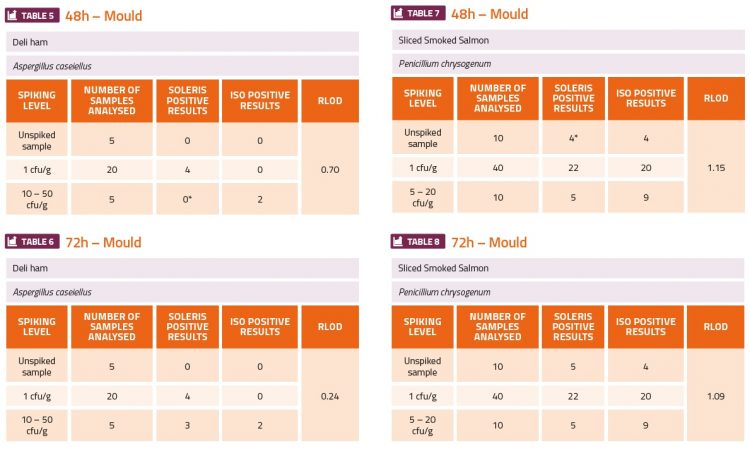
The challenge experiments used high (5-50cfu·ml-1 or cfu·g-1) and low (1cfu·ml-1 or cfu·g-1) levels of the yeast and mould organisms to contaminate fruit juice, tomato ketchup sauce, fruit yoghurt, deli ham, smoked salmon and tap water (environmental). The ranges of cfu indicated in the tables below were estimated from the plate counts which were done in parallel. The calculation of the relative level of detection (RLOD) between the two methods indicated which method was more sensitive. If the RLOD value was below 1 this would indicate that the Soleris method was more sensitive than the ISO plating method. If the value was between 1 and 1.5 this would indicate that both methods have a similar sensitivity. If the value was above 1.5, then this would indicate that the ISO plating method was more sensitive. The results shown in the tables below show that the Soleris system was more sensitive than the ISO 21527-1 method for all the yeast contaminants (Tables 1-4) and A. caseiellus (Table 5-6), and at least as sensitive as the ISO method for P. chrysogenum (Table 7-8). Both methods performed better at detection of yeast contamination than mould contamination, but there were issues with the dilution series for both A. caseiellus and P. chrysogenum since both organisms grew slowly on the potato dextrose broth and it was diffi cult to achieve a homogenous suspension for the dilution series, resulting in less accurate spiking levels. It was also evident from both methods that the smoked salmon sample was already contaminated with a fungal species.

Exclusivity test results
Bacterial species Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli, Enterococcus faecalis and Lactobacillus brevis were directly added to the vials to determine whether the presence of bacteria would give false positives in the system. The tests were carried out with levels likely to be found as contamination levels in food and beverage matrices (10-4, 10-5 and 10-6 dilutions of a seed culture (at stationary phase) were used) and they gave negative results. The exception was P. aeruginosa at a 10-6 dilution, which showed a positive result in this test. However, the 10-4 and 10-5 dilutions showed negative results, which suggests that the 10-6 dilution of P. aeruginosa was probably contaminated with fungi during the dilution series or contaminated whilst adding samples to the vials. It should be noted that these experiments indicated that a shuteye setting of ‘50’ was not suffi cient. When the shuteye was adjusted to 120 the initial drift in optical density units after introducing the vials to the machine was omitted from the readings and false positives avoided; the resulting optical density curves generated are the same, but the results were interpreted diff erently. These exclusivity tests showed that the Soleris system is likely to only give positive results for fungal organisms, as long as the bacterial concentrations are at levels likely to be encountered in foodstuff s and beverages and the settings on the machine are fi t for food matrix testing.
Discussion and conclusion
In this study, the Soleris Yeast and Mould system was shown to detect C. krusei, C. albicans, C. tropicalis, S. cerevisiae, A. caseiellus and P. chrysogenum in a variety of food and beverage matrices. The minimum concentration of contamination detected was 1cfu·ml-1. For the yeasts, the detection rate was very high (see Table 1-4). The spiking levels in the mould detection assay were compromised by the difficulties encountered in separating the mould clusters and obtaining an even suspension of the mould, but it also appeared to be more difficult for mould contaminants to reach the growth rate that triggered positive results. Moulds are known to be slow growers7, compared to yeasts8 so this probably accounts for the comparably lower levels of detection, but clumping of the moulds in the broth is also likely to have contributed. However, taking into account only results that showed a positive result within 48h, the Soleris system was shown to be more sensitive, or at least as sensitive, as the ISO standard for mould detection. In some cases, extending the test time up to 72h may lead to an improved sensitivity.

References
1. Pitt JI, Hocking AD (2009) Fungi and Food Spoilage 3rd edition, Dordrecht : Springer, Springer Dordrecht Heidelberg London, New York
2. WHO (2015) Fact sheet: food safety [online], available: www.who.int/mediacentre/ factsheets/fs399/en/[accessed 4th august 2016]
3. Groll, A.H. (2007) Introduction to Current Fungal Infection Reports, Current Fungal Infection Reports 1:1-2
4. Shelef LA, Firstenberg-Eden R (1997) Novel selective and non-selective optical detection of micro-organisms, Letters in Applied Microbiology 25: 202-206
5. Firstenberg-Eden R, Foti DL, Mcdougal ST, Baker J (2002) Optical instrument for the rapid detection of microorganisms in dairy products, International Dairy Journal 12: 225-232
6. Alles S, Shrestha N, Ellsworth A, Rider A, Foti D, Knickerbocker J, Mozola M (2009) Validation of the Soleris yeast and mould test for semiquantitative determination of yeast and mould in selected foods. Performance tested methods 040901, Journal of AOAC International 92:1396-1415
7. Meletiadis, J., Meis, J. F. G. M., Mouton, J. W., & Verweij, P. E. (2001) Analysis of Growth Characteristics of Filamentous Fungi in Different Nutrient Media. Journal of Clinical Microbiology, 39(2): 478–484
8. Klepser, M. E., E. J. Ernst, R. E. Lewis, M. E. Ernst, and M. A. Pfaller (1998) Influence of test conditions on antifungal time-kill curve results: proposed for standardised methods. Antimicrobial Agents & Chemotherapy 42:1207-1212
Issue
Related topics
Contaminants, Food Safety, Hygiene, Lab techniques, Quality analysis & quality control (QA/QC)









