Natural resistance in bacteria to biocides
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 13 December 2016 | Veslemøy Andersen (Global Harmonization Initiative) | No comments yet
Veslemøy Andersen, Global Harmonization Initiative (GHI), offers an insight into the natural resistance in bacteria to biocides…

The food industry uses biocides to prevent pathogenic microbes contaminating equipment as well as the food being processed. There is strong evidence to suggest that biocide resistance and antibiotic resistance in bacteria may be linked. This is called biocide-antibiotic cross-resistance or cross-tolerance.
An important issue of concern today is the possibility that pathogenic bacteria could develop resistance against biocides and pass this competence, including the possibility of multi-resistance, along the food chain and in this way further the development and spreading of antibiotic resistance in bacteria. Resistance mechanisms in bacteria can be divided into natural mechanisms (permeability barrier and biofilm formation) and acquired resistance mechanisms (mutations, acquisition of genes, impermeability, target site alterations, enzymatic transformation and efflux pumps). This article will focus on the two natural resistance mechanisms; permeability barrier and biofilm formation.
Permeability barrier
One of the most important natural resistance mechanisms is reduced susceptibility due to the chemical structure and composition of the outer cell layers of vegetative bacteria and bacterial endospores. This natural resistance is a chromosomal controlled character of the bacterial cell. The ability to withstand or resist an antimicrobial attack, due to this natural resistance, varies between different bacteria and sometimes also between various strains of the same bacterial species. These strains can become very tolerant to antimicrobials and withstand higher antimicrobial concentrations than is needed for killing most of the bacteria of the same species.
The most susceptible Gram-positive bacteria have no extra layers outside their peptidoglycan layer that can protect them from antimicrobial attack. Small molecule biocides, such as alcohols, aldehydes, phenols, QACs and biguanides have no problem penetrating the Gram-positive cell wall, which makes them the most sensitive of the bacteria. Gram-negative bacteria, however, have an outer membrane layer composed of a lipid bilayer and a lipopolysaccharide (LPS) layer outside the outer membrane protecting them from outside influence, making them less susceptible. They can also regulate the permeability through decreasing the synthesis of porins, the proteins that are involved in the uptake of biocides and antibiotics1.
The most resistant Mycobacteria have a complex cell wall outside their peptidoglycanpolysaccharide layer – a waxy layer and a lipid layer – making them the least susceptible of the vegetative bacteria. It is the mycoylacylarabinogalactan and arabinogalactan/arabinomannan in the cell wall that makes it almost impossible for biocides to reach the target sites in Mycobacteria2 . The bacterial endospores of Bacillales and Clostridiales (Figure 1) species are far more resistant to antimicrobials than all vegetative bacteria. Their spore wall is composed of layers that differ from the vegetative cell, which protect them against biocide action3 . The outermost layer, the exosporium, is a protein layer that protects the spore from antimicrobials and enzymes and forms the first defence. The outer membrane layer (outside the cortex) does not play a role in the permeability barrier. A peptidoglycan-containing germ cell wall surrounds the plasma membrane.
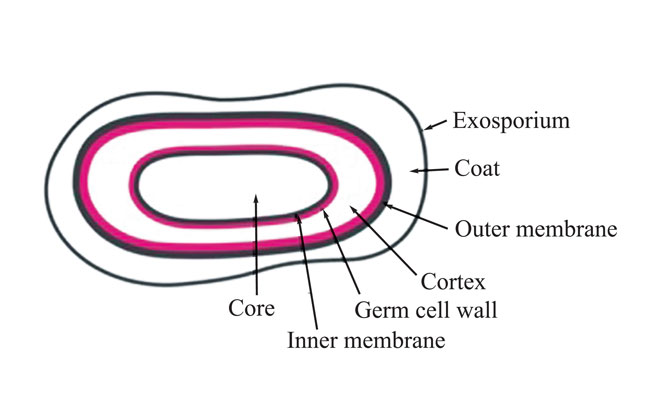

Figure 1: Bacterial endospore (E.V.M. Andersen, 2016)
The plasma membrane containing immobilised lipids forms a permeability barrier against small molecules, protecting the DNA, RNA and enzymes in the core. This lipid layer is thought to be the reason that the plasma membrane is so effective in protecting the spore core DNA from damage by biocides. Microorganisms other than bacteria and bacterial endospores – the fungi and enveloped viruses – are more susceptible to biocides than Gram-positive bacteria, the enveloped viruses being the most susceptible.
Biofilm formation
Another important form of natural resistance in bacteria is the formation of biofilms. Bacteria naturally prefer to live together in close contact with each other and not as planktonic (single cell or free floating) individuals. Biofilms are highly organised communities of bacteria cohabiting in extracellular polymeric substances (EPS) slime, consisting of polysaccharides, protein, lipids, phospholipids, nucleic acids and humid substances4,5; of which protein and polysaccharide accounts for 75-89 % of the slime composition6 . The EPS protects the biofilm community against threats from outside, such as biocides. It has been found that bacteria in biofilms are less susceptible to antibiotics and biocides.
The position of the bacteria in a biofilm reflects their susceptibility to biocides; having the most susceptible strains living underneath and the least susceptible strains on the top. The biocide concentration underneath is sub-optimal and might select for resistance in the bacteria living there. Biofilms that have the chance to stick firmly to the surface are more resistant to biocide attack than biofilms left there for only a few hours. Bacteria inside a steady-state growth biofilm can withstand biocide concentrations that are many times greater than their planktonic ‘sisters’ of the same strain. Reduced susceptibility to Triclosan has been found in Salmonella and in Proteus/Providentia7,8. In Enterobacter sakasakii an increased survival was found after exposure to QACs9 ; resistance to peroxides of Listeria in biofilms was found after their exposure to them10.
A biofilm is built up with advanced architectural structures with a tangled system of nutrient supply, water supply and waste matter drainage systems, with bacteria having their own way of communicating (Figure 2). This communication system – a self–organisation and regulation system of the bacterial cells – facilitates adaption to changing environments through modulation of gene expression11. It is called ‘quorum sensing’ and makes it possible for bacteria to communicate with each other through small diffusible organic signal molecules, called auto-inducers (AI), that are excreted and accumulate around the cells during growth. If the cell concentration is high, this results in a sufficient concentration of the signal molecule to start the expression of genes that are involved in biofilm development, especially attachment and detachment. The bacteria are using a different ‘language’ (another signal molecule) between bacteria of their own species, than between other bacteria of different species.
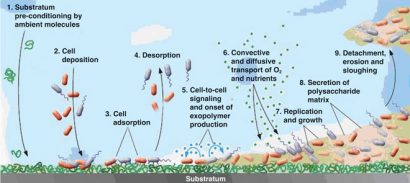

Figure 2: Biofilm formation steps. Reprinted with permission from the American Society for Microbiology (Breyers & Ratner, 2004. ASM News, Volume 70, Number 5, p. 232-235.
In Gram-negative bacteria this signal molecule is an N-acyl homoserine lactone molecule. The difference between different species is the length of the N-acyl homoserine part of the signal molecule.
In Gram-positive bacteria the signal molecules are oligopeptides. These biofilms can harbour single- or multi-species bacteria, in a single layer or three-dimensional structures. There are many different biofilms and under different environmental conditions (different food productions) a single type of bacteria can make different kinds. The steps in the formation of biofilms are as follows:
- Pre-conditioning. (2, 1). The attachment site is prepared with macromolecules existing in the liquid or intentionally produced. This alters the physiochemical condition of the surface (changes in hydrophobicity, electro – static charges, surface free energy)12.
- Desorption (2, 4). At this stage the bacteria are reversibly adsorbed to the surface and can detach.
- Immobilisation (2, 2 and 3). Deposition of planktonic bacteria cells on the surface and irreversible adsorption of these cells to the surface.
- Colonisation (2, 6 and 7). Nutrients and oxygen are transported to the various biofilm layers. The metabolic processes, DNA replication and cell growth of the bacteria in the biofilm begin.
- Consolidation (2, 5 and 8). When the concentration of bacteria is high enough, the bacterial cell-to-cell signalling system (quorum sensing) starts5 . EPS is formed, causing the bacterial cells to stick together and adhere more firmly to the substrate. The chemical structure of EPS usually varies among different types of bacteria and different environmental conditions13. The presence of extracellular filamentous elements on the bacteria cells, such as fimbria, pili and flagella, play an important role in the structure of the EPS. In Gramnegative bacteria, amyloid-like curly fibres together with these extracellular structures form the core architecture of the biofilm14. The upper layer consists of more curly fibres than the underlying layers, keeping the bacteria encased in a complex network of fibres and protecting the bacteria from outside stress factors, which makes them difficult to eradicate. In the underlying layers the presence of flagella is dominating14.
- Detachment (2, 9). Parts of the biofilm detach in order to propagate15. Biofilm detachment can be chosen for biological reasons. If, for example, bacteria in the biofilm experience their nutrient supply diminishing and too many of their own species are on the site where they live, then they can choose to leave the biofilm alone or together as a piece of the biofilm. It is a bit difficult to imagine that it is possible to leave the sticky, firm, slime matrix, but this is accomplished through sloughing steps where single cells detach and swim away with their flagella, or pieces of the biofilm are detached. The bacteria can also initiate this detachment step in the biofilm formation on their own. It has been found that Pseudomonas aeruginosa has an enzyme, alginate lyase, that can hydrolyse the slime matrix. This explains why bacteria, normally living in the inner part of the slime, can manage to rise to the upper parts of the biofilm to leave. Plank – tonic cells or bacteria in detached biofilm parts can leave and initiate the formation of a new biofilm elsewhere, which is called dispersal.
Conclusions
As of yet there are no useful techniques that can prevent biofilm formation without having unwanted side effects. Attachment of the planktonic bacteria to the surface is the basis of a successful biofilm formation. The best strategy is to thoroughly clean and disinfect the equip – ment in time such that bacteria does not have the chance to firmly attach to the surfaces. Cracks on the surfaces of equipment must also be avoided, because it makes it difficult to clean and therefore can be a place where biofilm formation starts.
An important factor in biofilm formation is the bacterial communication system, quorum sensing, that makes it possible for them to keep in contact with each other and communicate about every important issue in the development and maintenance of these ‘biofilm cities’ of bacteria. In the race to make new antibiotics, much work is carried out with aim of finding antiquorum sensing molecules that stop the bacteria from communicating. If it is possible to use these antiquorum molecules to stop biofilm formation, it will be easier to keep the equipment free from biofilms. This will make it possible to destroy all bacteria during the disinfection treatment and thus eliminate the problem of selection of resistant bacteria, resulting from the not fully effective use of biocides.
More detailed information on Bacterial resistance to biocides can be found in Andersen (2016)16.
About the Author
Veslemøy Andersen has a PhD degree in general microbiology from the University of Oslo, Norway. Her career began as a microbiologist/researcher at The National Institute of Public Health, Oslo. After a postdoc at the University of Groningen, The Netherlands, she worked at the University of Applied Sciences in Eindhoven, The Netherlands, setting up the curriculum of a new study programme ‘Food Microbiology’. Currently she is working for the Global Harmonization Initiative (GHI), Vienna, Austria, as a microbiologist and editor of the new GHI book series on Nutrition and Health Aspects of Traditional and Ethnic Foods.
References
- Poole, K., 2002. Mechanisms of bacterial biocide and antibiotic resistance. Journal of Applied Microbiology 92 Suppl, 55S-64S
- Anderson, G.G., O’Toole, G.A., 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiology Immunology 322, 85-105
- Leggett, M.J., McDonnell, G., Denyer, S.P., Setlow, P., Maillard, J.Y., 2012. Bacterial spore structures and their protective role in biocide resistance. Journal of Applied Microbiology 113, 485-498
- Jahn, A., Nielsen, P.H., 1998. Cellbiomass and exopolymer composition in sewer biofilms. Water Science and Technology 37(1), 17-24
- Sutherland, I., 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3-9
- Tsuneda, S., Aikawa, H., Hayashi, H., Yuasa, A., Hirata, A., 2003. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiology Letters 223, 287-292
- Stickler, D.J., Jones, G.L., 2008. Reduced Susceptibility of Proteus mirabilis to triclosan. Antimicrobial Agents and Chemotherapy 52, 991-994
- Williams, G.J., Stickler, D.J., 2008. Some observations on the migration of Proteus mirabilis and other urinary tract pathogens over foley catheters. Infection Control and Hospital Epidemiology 29, 443-445
- Kim, H., Ryu, J.H., Beuchat, L.R., 2007. Effectiveness of disinfectants in killing Enterobacter sakazakii in suspension, dried on the surface of stainless steel, and in a biofilm. Applied Environmental Microbiology 73, 1256-1265
- Pan, Y., Breidt, F., Jr., Kathariou, S., 2006. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Applied and Environmental Microbiology 72, 7711-7717
- Daniels, R., Vanderleyden, J., Michiels, J., 2004. Quorum sensing and swarming migration in bacteria. FEMS. Microbiol. Rev. 28, 261-289
- Dickson, J.S., Koohmaraie, M., 1989. Cell surface charge characteristics and their relationship to bacterial attachment to meat surfaces. Applied and Environmental Microbiology 55, 832-836
- Momba, M.N.B., Kfir, R., Venter, S.N., Cloete, T.E., 2000. An overview of biofilm formation in distribution systems and its impact on the detoriation of water quality. Water SA 26, 59-66
- Serra, D.O., Richter, A.M., Hengge, R., 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. Journal of Bacteriol 195, 5540-5554
- Breyers, J.D., Ratner, J.P., 2004. Bioinspired implant materials befuddle bacteria. American Society for Microbiology News 70, 232-237
- Andersen, E.V.M., 2016. Chapter 39. Bacterial resistance to biocides. In: Handbook of hygiene control in the food industry, 2nd ed., Lelieveld, H.L.M., Holah, J. and Gabric, D. (Eds). Woodhead Publishing Limited (imprint of Elsevier), Langford Lane, Kidlington, UK