Yeast forensics: methods for identification and tracking
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 3 December 2008 | Dr Suzanne Jordan, Molecular Methods Manager, Campden BRI | No comments yet
Yeasts are a diverse range of organisms, many of which are beneficial to the food and drinks industry in fermentation and the flavour development of dairy, meat and beverage products. There are also strains that spoil products, resulting either in blowing packs or off odours and flavours.
Yeasts are a diverse range of organisms, many of which are beneficial to the food and drinks industry in fermentation and the flavour development of dairy, meat and beverage products. There are also strains that spoil products, resulting either in blowing packs or off odours and flavours.
Yeasts are a diverse range of organisms, many of which are beneficial to the food and drinks industry in fermentation and the flavour development of dairy, meat and beverage products. There are also strains that spoil products, resulting either in blowing packs or off odours and flavours.
It is difficult to approximate the financial losses to the industry caused by spoilage, and there are no published figures available for the United Kingdom. In Australia, the calculated losses due to fungal spoilage are reported to be AUD 10,000,000 per annum1. Recent reports indicate that 20 per cent of UK domestic waste taken to landfill was spoiled, unused or leftover food2.
A range of techniques are available to assist the industry in answering questions, such as identifying spoilage organisms, determining culture authenticity and tracking sources of contamination. Identification provides a name for an unknown organism to either genus or species level and can be used to search for information on spoilage or safety issues and highlight the potential for resistance to preservation strategies. Characterisation allows organisms that share similar characteristics, such as DNA fragment profile, to be placed into groups, which assists in determining potential sources of contamination. These techniques can be divided into two groups based on their approach, i.e. biochemical and molecular based assays.
Biochemical tests rely on the fact that different types of yeast will vary in the sugars they are able ferment and the by-products they produce. These variations can be detected through a change in either colour or turbidity. Traditional schemes typically used a combination of physiology and fermentation tests to identify isolates3,4.
Further improvements in biochemical based methods have seen the introduction of selective media that enables differentiation of some medical isolates of Candida spp.5, and the development of miniaturised identification test panels. These panels usually contain dehydrated reagents to assist in test preparation. The end point reactions are read to create a ‘profile’ which can be searched against the reference database (held on a disc or web server). There are numerous assays available including the RapID system (REMEL), API 32C, API 20C and Aux system (bioMerieux), Auxacolour (Sanofi, Paris) and Yeast Star (Clarc Laboratories, the Netherlands). More recent developments in biochemical based methods include the automation of sample preparation and reading of test results, which reduces both hands-on time and the subjectivity when interpreting results. An additional advantage of the automated systems is the shorter time to result through assay optimisation. Examples of such systems include the VITEK 2 compact (bioMérieux) shown in Figure 1.


Molecular tests analyse either the genetic ‘code’ of the organism or the bar-code like fragment profiles resulting from cleavage of DNA (restriction digests) or the selective amplification of targets (during Polymerase Chain Reaction, PCR). Amongst the most commonly used techniques are DNA sequence analysis, DNA microarrays and PCR based assays. There is an increasing recognition of the benefits of such techniques by leaders in the field of microbiology as well as regulatory bodies. In their guidelines to the pharmaceutical industry for the aseptic processing of sterile drug products (using current good manufacturing practice), the Food and Drug Administration (FDA) states that molecular (genotypic) methods have been shown to be more accurate and precise than traditional biochemical and phenotypic techniques6.
Most of the molecular based identification is done by sequence analysis of conserved targets such as ribosomal DNA (18S or 26S rDNA) which provides a relatively stable point of reference. Ribosomal DNA has a relatively slow evolutionary rate which lends itself to applications such as microbial identification. Additional differentiation can be achieved using regions that show a greater degree of variability between species such as the internal transcribed spacer (ITS). Sequence analysis is a multi stage technique that determines the order of the genetic ‘code’ (bases designated A, C, G and T) of a target fragment amplified (by PCR) from an unknown organism. This data is compared against a reference database to give a percentage match. A typical workflow for sequence analysis is shown in Figure 2 overleaf. This technology has the capability to provide both rapid and definitive answers to the identification of unknown isolates. The benefits of sequence based identification include independence on growth or fermentation, which are the limiting factors for biochemical tests. An illustration of this was observed in a case study where a yeast (with an atypical biochemical profile) isolated from a dairy product was identified by sequence analysis (Microseq, Applied Biosystems) as Cryptococcus sp. Another example involved osmophilic yeast (from a high water activity product) which was identified as Zygosaccharomyces rouxii.
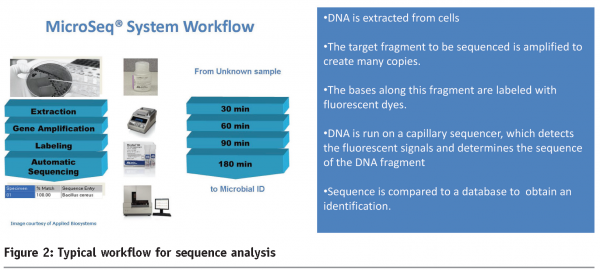

The availability of ‘off the shelf’ systems for sequence based identification has provided further benefits, including standardisation (of reagents and protocols), inbuilt quality thresholds (for automated data analysis) and the availability of validated databases and software which adds to the confidence of the results generated. Future areas of development may include using multiple targets to improve discrimination between closely related species. Multi locus sequence typing (MLST) is well developed for bacterial isolates7 and has shown potential in the rapid identification of pathogenic moulds8.
In addition to identification, DNA assays have been developed for the characterisation of yeast isolates. A variety of methods have been used for characterisation, including amplified fragment length polymorphism (AFLP), PCR restriction fragment length polymorphism (RFLP), random amplified polymorphism detection (RAPD), pulsed field gel electrophoresis (PFGE), MLST, variable number tandem repeats (VNTR) and repeat based PCR (rep PCR). The majority of these techniques were initially used for bacterial isolates and have been adapted for analysis of yeast isolates. PFGE is recognised as the gold standard approach for the characterisation of bacterial pathogens (particularly for epidemiological investigations) and to assist rapid dissemination of data. International and national databases have been established such as EnterNet (Funded by the European Commission – DG SANCO) and PulseNet (developed by CDC in association with the Association of Public Health Laboratories APHL) respectively. For yeast isolates, the method is often referred to as karyotyping and has been used to analyse isolates in many food sectors including brewing strains within the alcoholic beverage industry. Although this technique can be highly informative, it does require a relatively high level of skill to achieve consistent results. Repeat based PCR (rep PCR) targets signature regions that occur along the genetic code which are amplified during PCR. These are separated (resolved) to create a unique fingerprint that can be compared to other patterns to enable their similarity to be assessed.
The technique is able to discriminate between isolates of Saccharomyces cerevisiae, with subtle differences in profile. Analysis of the patterns revealed that the lager yeasts (NCYC 1056 and NCYC 1073) clustered together at 99.6 per cent similarity. Although the ale and wild type yeasts (NCYC 1045 and BRX 367 respectively) share some similarity between profiles with each other and the lager yeasts, they each possess unique characteristic patterns. A reference isolate Candida utilis has been included to put the similarities into context. The availability of a standardised automated rep-PCR system (using lab on a chip technology) will assist in the adoption of the technique, particularly where high throughput and rapid time to result analysis (i.e. same day) is required.
The field of microbial identification and characterisation has evolved dramatically and now has the potential to provide same-day answers for industry. This is often key in the effective management of contamination and spoilage issues. Additional benefits of the new methods include the potential for high throughput and reduced subjectivity during interpretation of results. At Campden BRI, we have a range of techniques available for identification and characterisation of yeast isolates using biochemical and molecular based methods. These technologies assist us in providing timely definitive answers to enable our clients to respond rapidly to food safety and spoilage incidents.
References
- Pitt J.I. and A.D. Hocking (1997) In Fungi and food spoilage, 2nd edition. Blackie Academic and Professional, London
- Anon (2007) In Beyond packaging: food waste in the home. IGD, Hertfordshire, U.K
- Deak T. and L. R. Beuchat (1987). Identification of foodborne yeasts. J. Food Prot. 50: 243-264
- Barnett J.A., R.W. Payne and D. Yarrow (2000) In Yeasts: characteristics and identification. 3rd Ed. Cambridge University Press, Cambridge, England
- Eraso E., Moragues M.D., Villar-Vidal M., Sahand I.H., González-Gómez N., Pontón J., Quindós G. (2006) Evaluation of the new chromogenic medium Candida ID 2 for isolation and identification of Candida albicans and other medically important Candida species. J. Clin. Microbiol. 44:3340-5
- Anon (2004) Guidance for Industry Sterile Drug Products Produced by Aseptic Processing – Current Good Manufacturing Practice. Pharmaceutical CGMPs. Food and Drug Administration, Rockville, USA
- Maiden M.C., J.A. Bygraves, E. Feil, G. Morelli, J.E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D.A. Caugant, I.M. Feavers, M. Achtman, and B.G. Spratt. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 95:3140-5
- Rakeman J.L., U. Bui, K. LaFe, Y-C. Chen, R.J. Honeycutt, and B.T. Cookson (2005) Multilocus DNA sequence comparisons rapidly identify pathogenic moulds. J. Clin. Microbiol. 43: 3324-3333









