Use of molecular techniques in the food industry
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 1 June 2009 | Mieke Uyttendaele and Andreja Rajkovic, Laboratory of Food Microbiology and Food Preservation, Ghent University | 1 comment
Microbial analysis in foods is an integrated part of management of microbial safety in the food chain. Both competent authorities and individual food business operators use microbial analysis for monitoring of the actual situation and trend analysis in order to detect emerging risks. For compliance testing to defined microbiological criteria or assessment of the performance of management strategies based upon HACCP, microbial analysis is also a valuable tool. Molecular techniques, especially the polymerase chain reaction (PCR), are one of the most important rapid methods for the sensitive and specific detection of pathogenic micro-organisms.
Microbial analysis in foods is an integrated part of management of microbial safety in the food chain. Both competent authorities and individual food business operators use microbial analysis for monitoring of the actual situation and trend analysis in order to detect emerging risks. For compliance testing to defined microbiological criteria or assessment of the performance of management strategies based upon HACCP, microbial analysis is also a valuable tool. Molecular techniques, especially the polymerase chain reaction (PCR), are one of the most important rapid methods for the sensitive and specific detection of pathogenic micro-organisms.
Microbial analysis in foods is an integrated part of management of microbial safety in the food chain. Both competent authorities and individual food business operators use microbial analysis for monitoring of the actual situation and trend analysis in order to detect emerging risks. For compliance testing to defined microbiological criteria or assessment of the performance of management strategies based upon HACCP, microbial analysis is also a valuable tool. Molecular techniques, especially the polymerase chain reaction (PCR), are one of the most important rapid methods for the sensitive and specific detection of pathogenic micro-organisms.
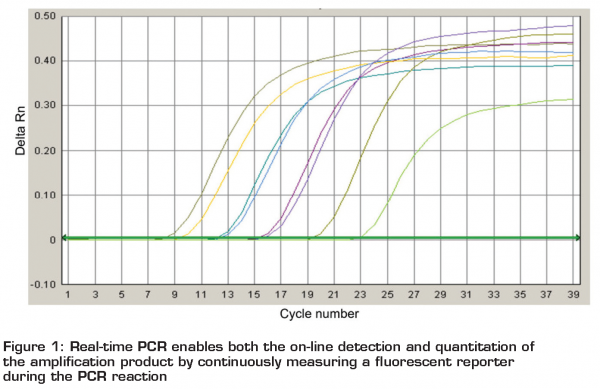
The PCR technique, first described by Kary Mullis in the mid-1980s is a three-step cyclic in vitro procedure based on the ability of the DNA polymerase to copy a strand of DNA. When two primers bind to complementary strands of target DNA, the sequence in between is amplified exponentially with each cycle making the technique a very sensitive tool. The presence of even one copy of the template within the reaction mixture can be detected within a couple of hours as approximately a million-fold of copies are created. The results of PCR are traditionally (conventional PCR) detected by agarose gel electrophoreses and staining and specificity of the bands may be further identified by sequencing. In the early 1990’s, the ‘second’ generation of PCR technologies was introduced by the use of fluorescent double stranded DNA dyes e.g. SYBR@ Green or DNA probes e.g. Molecular beacons® or TaqMan® probes. Real-time PCR enables both the on-line detection (Figure 1) and quantification of the amplification product signal by continuously measuring a fluorescent reporter during the PCR reaction. The potential application of PCR also in routine labs was boosted not only the fact that PCR is no longer a black box reaction but also the fact that real-time PCR consists of a closed-tube format, reducing the risk of contamination potentially leading to false-positive results.
Sample preparation, a prerequisite to reliable PCR detection of pathogens in the food lab
The terminology ‘rapid’ and ‘sensitive’ in the description of the PCR technique as a detection method for food borne pathogens is relative. PCR as such takes only approximately 30 – 90 minutes. However, it detects genomic DNA copies in a PCR tube, rather than bacterial cells in foods as requested in legal criteria. For detection of low numbers i.e. one to 10 cells per 25 grams, thus a prior cultural enrichment procedure is normally applied. Overall, for reliable PCR detection, the low numbers of cells (approximately one in 10 grams of food sample) need to grow during prior enrichment to levels of at least 100 cells per millilitre of the primary suspension. Present PCR-based validated proprietary methods on the market for detection of food borne pathogens indeed recommended the enrichment step six to eight hours up to 24 hours prior before proceeding to DNA extraction and execution of PCR. Shortened enrichment procedures, overlooking the need to repair stressed and/or sub-lethal injured cells may result in a true risk of not reaching the detection limit of the subsequent PCR technique and can lead to false negative results. If a further reduction of time to detection is needed, further progress on elaboration of sample preparations procedures should be made. Focus is actually put on concentration protocols of bacteria or DNA from larger volumes e.g. the use of bacteriophage-based capture, the use of nanoparticles, devices enabling upscaling of present available capture or extraction methods, etc. Nevertheless, in the concentration of cells (or DNA), care has to be taken not to concentrate potential interfering factors from the food matrix as well, as this will jeopardise the efficient functioning of the PCR reaction which is prone to inhibition and in turn might lead to false negative results.
PCR, nowadays an accepted method in the food lab
PCR has evolved to a convenient and rapid method in recent years. The further standardisation of PCR protocols, a convenient PCR kit format and elaboration of user-friendly software interfaces for interpretation of PCR results especially enabled the use of the PCR technique also by non molecular biology trained lab technicians and has increased the accessibility of PCR to many routine labs. In addition, in the meantime, the suppliers of PCR reagents or kits have built up a strong customer support system, offering a wide range of tailored PCR products and having acknowledged the particular challenges of PCR as a tool in food microbiology. Optimisation of PCR has made it an intrinsic extremely sensitive technique and DNA is a very stable (and thus amplifiable) molecule. Aerosols created during operation of PCR may bring DNA fragments in the lab environment (and subsequently in the PCR tube). Therefore, especially positive results with Ct values > 40 (overall indication of very low levels of DNA near the theoretical detection limit) should be interpreted with care. Real-time PCR is a rapid tool for screening of samples, still in case of positive PCR results it could be tempting to confirm the positive result by means of the culture-based method. However, in particular cases the culture method may not have proven selectivity to detect the pathogen and provide an isolate. Nowadays, after 20 years of introduction of PCR in the lab, food microbiologists have lost their initial hesitation to this ‘high tech’ PCR technique and accepted that PCR is a complementary tool to the set of rapid diagnostic test methods available in microbial analysis in foods. Progressive introduction of automated and/or miniaturised equipment for PCR has enabled it to become a high through-put method. As such, PCR methods are increasingly applied in surveillance and monitoring programs both with competent authorities and in food industry to detect pathogens such as Salmonella, Listeria monocytogenes and E. coli O157 in a variety of samples including food and feeding stuffs, veterinary and environmental samples
Ongoing developments
Multifunctional approach
In diagnostic labs, there is a continuous search for multifunctional and convenient formats to elaborate PCR in order to get the maximum information in the minimum amount of time and cost. A recent innovation in this field is the introduction of the Genesystems® PCR technology. The GeneDisc is a disposable plastic reaction tray, the size of a compact disc. Its rim is engraved with 36 reaction microchambers preloaded with dessicated PCR reagents necessary for detection of the required targets. For example, a GeneDisc format exists divided up in six chambers allowing parallel examination of six samples of presumptive O157 EHEC isolates for simultaneous detection of Shiga toxins 1 and 2 (stx1 and stx2), intimins (eae), the somatic O157 associated gene and the flagellar H7 gene. A second GeneDisc exists for the identification of predominant EHEC serotypes by detection of O-group-associated genes of EHEC O26, O103, O111, O145 and O1571.
Quantitative PCR (Q-PCR) enabling enumeration of food borne pathogens
Development of real-time PCR has enabled a quantitative approach. However, in order to keep the relation between the numbers of bacteria in the food and the enumeration by PCR, a prior enrichment step is prohibited. Therefore, there is a need for a dedicated (often labour-intensive) sample preparation for recuperation of cells from the food and acquiring good quality DNA for amplification. The bottleneck for enumeration of bacterial cells by Q-PCR is thus the sample preparation. In addition, additional process controls will have to be taken along to check out on the potential loss of target bacteria during the extraction procedures. At present, enumeration of L. monocytogenes in smoked salmon by Q-PCR is reported2. However, the limit of quantification is situated at approximately 103-104 cells per gram. This limit is still too high for practical application, since most samples taken in along the food chain are contaminated with lower numbers of pathogens (usually less than 100/g).
Detection of Norovirus by reverse transcriptase PCR (RT-PCR)
Norovirus is the predominant food borne virus. The human diarrheal Norovirus strains are at present non-culturable in established cell lines. Thus, detection in case of food borne outbreaks or monitoring programs relies on RT-PCR as Norovirus is an RNA-virus. Because of the lack of culturability, direct extraction of virus particles and/or extraction of viral RNA from suspected food are a necessity with associated difficulties in developing reproducible and reliable extraction procedures. Up to now, there is no standard method available for RT-PCR detection of Norovirus in foods. The genetic variability of the virus troubles the selection of a universal target gene sequence Also, extraction of RNA sets (even more) challenges sample preparation as it is more easily degraded than DNA during extraction. In addition, no straightforward relation between detection of genomic copies and the presence of infectious virus particles could be established (Baert et al. 2008)3.
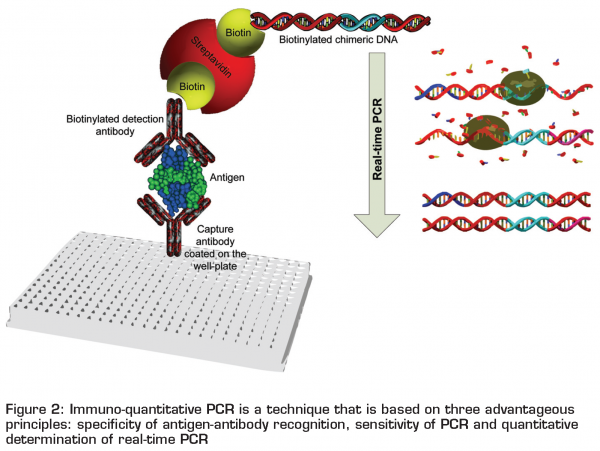
Immuno-quantitative PCR (iqPCR) for detection of bacterial toxins
Immuno-quantitative PCR is a technique that is based on three advantageous principles: specificity of antigen-antibody recognition, sensitivity of PCR and quantitative determination of real-time PCR through real-time monitoring of the exponential signal amplification. The principle of the technique is schematically presented in Figure 2 on page 24. Its main advantage to other immunological methods appears to be its high sensitivity. Rajkovic et al.4 reported sensitivity in detection of Staphylococcus aureus enterotoxins B of 10 pgml-1 with iqPCR, which was approximately 1,000 times higher than with the ELISA using the same antibodies. A similar increase in sensitivity was observed in conventional PCR in detection of Clostridium botulinum neurotoxin A.
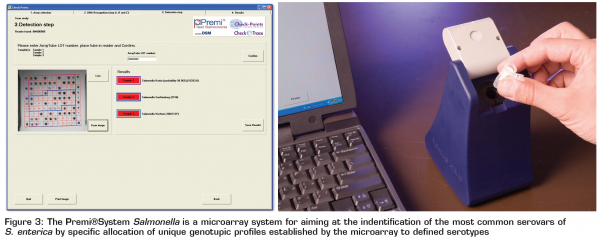
DNA microarrays in analysis of food borne bacteria
DNA microarrays are arrays of recognition ligands, such as oligonucleotide or cDNA molecules immobilised (chemically bonded) in discrete locations on a solid matrix. The target sequence to be analysed is labelled and hybridised to recognition probes on the array. The signal generated by the bound labelled target on the array allows identification based on the known locations of the probes. Different technologies for design and production of microarrays exist and the choice of a microarray technology depends on the details of the application. The most common application of microarray technology is differential gene expression analysis with the main objective being comparison of the expression levels of selected genes under control and test (often stress) conditions. Selected genes often cover functional, virulence or stress genes. Since DNA arrays allow simultaneous measurements of thousands of interactions between mRNA-derived target molecules and genome-derived probes, they are rapidly producing enormous amounts of raw data never before encountered by biologists. The bioinformatics solutions to problems associated with the analysis of data on this scale are a major current challenge. Next to this permanent issue of sample preparation and genetic material purification remains an issue for successful and reliable analysis. Next to this, detection of pathogens using microarrays has been described, as well as identification, genotyping and pathotyping (detection of appropriate virulence factors). Recently, a microarray system, the Premi®System Salmonella system (Figure 3) has been brought to the market by DSM Nutritional Products aiming at identification of the most common serovars of S. enterica. Starting from a presumptive Salmonella colony, results on the serotype/genotype of Salmonella are obtained within eight hours. The system allows single-tube processing of the samples and requires no peculiar technical skill. As such it shows interesting potential to be used for routine laboratory testing5.
Conclusion
PCR is increasingly applied in surveillance and monitoring programs. The scope of PCR techniques is continuously broadened and an increasing number of applications in food industry reported. The PCR technique now enables, in addition to the detection of practically all food borne pathogens, also detection of food borne viral agents such as the Norovirus via Reverse Transcriptase PCR (RT-PCR) and detection of bacterial toxins via immuno-PCR. In addition, the introduction of real-time PCR technology (RTi-PCR) enabling recording the increase of PCR product on-line rather than at the end of the PCR, allowed quantification of target DNA sequences and thus indirectly, also bacteria. Still, the break-through to routine application in food industry since the PCR technique was invented in the mid 1980s has taken some time. This can be attributed to challenges linked to complexity of the food matrix, often leading to interference of the in vitro enzymatic reaction. In addition, the small volumes used in a PCR reaction versus demands of food industry to detect low numbers of pathogens in quite voluminous amounts of food needs appropriate sample preparation. These bottlenecks have largely been overcome now and PCR techniques are readily accepted as being reliable detection methods for pathogens in the food industry. Already, more advanced molecular techniques such as micro-arrays are now popping up at the corner, finding its way to application in especially characterisation and identification of pathogenic isolates.


References
- L. Beutin, S. Jahn, P. Fach . 2009. Evaluation of the ‘GeneDisc’ real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. Journal of Applied Microbiology 106, 1122-1132
- Rodriguez-Lazaro et al. 2005. A novel real-time PCR for Listeria monocytogenes that monitors analytical performance via an internal amplification control. Applied and Environmental Microbiology 71, 9008-9012.
- L. Baert, C. Wobus, E. Van Coillie, L. Thackray, J. Debevere, M. Uyttendaele. 2008. Detection of Murine Norovirus 1 using plaque assay, transfection assay and real-time RT-PCR before and after heat exposure. Applied and Environmental Microbiology 74, 543-546
- A., Rajkovic, B. El Moulij, M. Uyttendaele, P. Brolet, W. Zorzi, E. Heinen, E. Foubert, J. Debevere 2006. Immuno-quantitative real-time PCR for detection and quantification of Staphylococcus aureus enterotoxin B in foods. Applied and Environmental Microbiology. 72, 6593-6599
- P. Wattiau, T. Weijers, P. Andreoli, C. Schliker, H. Vander Veken, H.M.E. Maas, A. J. Verbruggen, M.C. Heck, W. J. Wannet, H. Imberechts, P. Vos. 2008. Evaluation of the Premi®Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. International Journal of Food Microbiology 123, 293-298
Issue
Related topics
Food Safety, Polymerase Chain Reaction (PCR), Quality analysis & quality control (QA/QC)










We have learn alot
thank you