Whole Genome Sequencing and the food industry
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 8 November 2016 | Greg Jones, Senior Research Officer, Microbiology, Campden BRI | No comments yet
Whole Genome Sequencing (WGS) has the potential to render other forms of microbiological identification obsolete. New Food takes a closer look…


Figure 1: A small portion of a genome assembled from raw sequence reads. Each of the small black bars represents one sequence approximately 500 bases long.
Whole Genome Sequencing (WGS) has the potential to render other forms of microbiological identification obsolete. It is more accurate than a serotype, more discriminatory than a pulsed-field gel electrophoresis assay and it can prove relationships between strains with higher resolution than ever before. This is the method which has been adopted by regulatory agencies such as Public Health England and the Food and Drug Administration in the USA. Food companies are starting to become more aware of this area and wish to have a constructive dialogue with government agencies, however there can be a lack of knowledge regarding the technology and the potential uses of it in an industrial setting.


The technique has become more prevalent in the last five years due to rapid advances in sequencing technology that have led to dramatic falls in cost per sequence. It is now possible to sequence genomes on a routine basis for a few hundred pounds each. Regardless of the technique used, the generation of huge amounts of sequence data has become entirely unremarkable. When a genome is sequenced, the initial output is hundreds of thousands of short sequences. Each of these sequences is a few hundred bases long and represents a tiny fragment of the total genome. The next challenge is to assemble these reads by comparing them to each other and ordering them according to their overlapping ends. This process is analogous to reconstructing an ancient document from fragments of parchment.

Figure 1: A small portion of a genome assembled from raw sequence reads. Each of the small black bars represents one sequence approximately 500 bases long.
The workflow to assemble a genome is straightforward:
- Obtain an isolate via culture-based methods.
- Extract the DNA.
- Prepare the DNA for sequencing (“Library Preparation”).
- Run the sequencer.
- Assemble the short raw reads into longer sequences using software.
The next step is to give it some meaning by comparing it to other sequences. The comparison is reliant on the number of other genomes against which your submitted sequence is compared. The analysis gets larger as more genomes are added in to the comparison. Comparing whole genomes against databases of other whole genomes is currently performed by the FDA in the USA using their ‘Genome Trakr’ service. This service relies on the vast storage and computing power available to them from the National Centre for Biotechnology Information (NCBI). This is a resource available to anybody, and a genome submitted for analysis will be placed into context via comparison against other sequences. The output is a phylogenetic tree similar to the one shown in Figure 2.
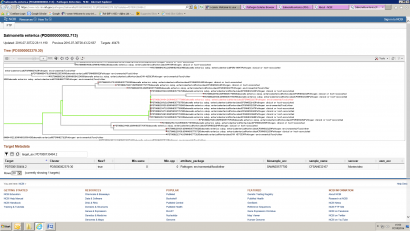

Figure 2: Output from Genome Trakr
In the example above, the genome labelled as an ‘Environment/Food’ sample is highlighted in red, and is shown to cluster very closely with a set of isolates designated as ‘Clinical’.
The necessary use of a public database for this analysis has led to concern from some in the food industry who fear that doing the right thing and submitting sequences will reflect badly on them in the event their sequence is shown to be related to an outbreak. Despite assurances from the FDA in a recent meeting, US industry representatives still had some concerns that samples submitted with accompanying descriptions of its source that could ultimately be traced back to the company of origin are so sensitive that in-house legal advice is to not submit at all. The mood in the UK is similar, with companies approaching this method with a degree of caution.
Is this caution warranted?
A similar tool for tracing outbreaks exists in the form of PulseNet, based on DNA fingerprinting technology. What is new for WGS is the finer level of discrimination. As this information is available to anyone, the submitting company will be alerted to any clinical link at the same time as the regulator, allowing earlier action to be taken. Submitters’ names are not made publicly available, but could be held by the regulator. Industry is therefore more likely to submit sequences if their describing metadata can be made anonymous. Earlier notification of a link to an outbreak is in everyone’s best interest, and it will be in the submitter’s interest to be removed from the investigative focus if the submitted sequence does not match clinical data. Despite these clear advantages, there is still the worry that a current isolate can be linked to outbreaks that occurred at any time, and that a current outbreak could be linked to an isolate submitted at any time. If the food industry is to use this technique and work constructively with the regulators, these issues need to be addressed and binding assurances given by the regulator that the industry’s desire to protect public health through the use of WGS will not result in an increased probability of prosecution should an unfounded link be made.
Campden BRI is actively working with industry and regulators to advise and help reach a mutually beneficial result. If you would like to explore Whole Genome Sequencing in more depth, please get in touch.









