Luminescent techniques for microbiological analysis of foods
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 7 March 2007 | Dr. Mansel Griffiths, Canadian Research Institute for Food Safety, Canada | No comments yet
There are many naturally bioluminescent organisms existing in nature and the mechanisms whereby some of these creatures emit light have been fully characterised1. These include the luciferin-luciferase system of bacteria, insects (fireflies and click-beetles) and the jellyfish Aequorea victoria. In essence, bioluminescence involves the conversion of chemical energy into light energy by an enzyme, commonly termed luciferase.
There are many naturally bioluminescent organisms existing in nature and the mechanisms whereby some of these creatures emit light have been fully characterised1. These include the luciferin-luciferase system of bacteria, insects (fireflies and click-beetles) and the jellyfish Aequorea victoria. In essence, bioluminescence involves the conversion of chemical energy into light energy by an enzyme, commonly termed luciferase.
There are many naturally bioluminescent organisms existing in nature and the mechanisms whereby some of these creatures emit light have been fully characterised1. These include the luciferin-luciferase system of bacteria, insects (fireflies and click-beetles) and the jellyfish Aequorea victoria. In essence, bioluminescence involves the conversion of chemical energy into light energy by an enzyme, commonly termed luciferase.
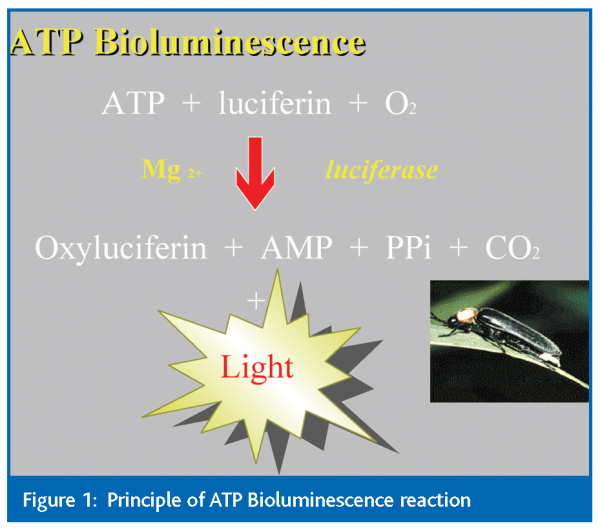
The luciferases from different organisms catalyse different reactions but all require oxygen. Over the years there has been great interest in developing applications based on bioluminescence2,3. A major advantage of using bioluminescent systems as analytical tools is that extremely low levels of enzyme activity can be detected by measuring the emitted light. Modern instruments are capable of detecting single photons with both temporal and spatial distribution; thus providing accurate information on the location and intensity of the light source4. Another feature of bioluminescent systems that makes them an excellent investigative tool is an almost absolute specificity for their substrates. For example, for firefly luciferase even minor changes in the structure of ATP and firefly luciferin result in a total loss of enzyme activity and this consequently results in a loss of light emission. This specificity for substrates allows the real-time measurement of luciferase activity in situ in very complex samples without the need for any pre-treatment.
ATP bioluminescence
The best known among bioluminescent methods is the ATP-assay, based on the activity of firefly luciferase (Figure 1). This assay has been used to determine biomass and is based on the principle that all living cells contain ATP and the ATP levels are proportional to the number of cells present. Thus, the amount of light emitted by the luciferase/luciferin reaction following extraction of ATP from cells provides an estimate of cell populations. This assay has been applied to estimate microbial load in a variety of foods including milk5-10, poultry11-14, meat 15, 16 and produce17. Results can be obtained within approximately 15 minutes and provide an accurate indication of whether counts have exceeded pre-determined thresholds. ATP bioluminescence has also been used to monitor the quality of processing waters during food manufacture 11, 18.
Arguably the most widely used application of this technology is ‘ATP Hygiene Monitoring’. This involves assessment of ATP levels on environmental swabs which directly relates to the cleanliness of the surface19, 20. Several commercial systems are available (Figure 2). Concern has been raised about inhibition of the luciferase in the presence of disinfectants remaining on surfaces following sanitation21-23. One elegant way to overcome this problem is to use mutant luciferases with increased resistance to the disinfectants used in industry24. In a twist to hygiene monitoring, it has been proposed that these tests can also be used to assess the potential for the presence of allergens on food contact surfaces.
Using methods for differential extraction of ATP from prokaryotic and eukaryotic cells it has been possible to develop assays for microbial cells that can be performed in minutes. However, the detection limit for these assays is high (>106 CFU/ml) and methods to improve their sensitivity have been a preoccupation of many microbiologists over the last two or three decades. To improve sensitivity, techniques involving filtration or centrifugation have been investigated and these have made it possible to detect approximately 104 CFU/ml in foods. This sensitivity can be further improved through assays of adenylate kinase, an enzyme present in cells that can be made to produce ATP by the addition of ADP in excess26. Another method to increase sensitivity involving the recycling of ATP by combining the enzyme pyruvate ortho-phosphate dikinase with luciferase/luciferin has been proposed by Sakakibara et al.27. These authors claim that their method is approximately 40 times more sensitive than that based on the luciferase/luciferin reaction alone.
Phage-based diagnostics
Another drawback of ATP bioluminescence assays is their inability to distinguish between the types of organisms present either in food or on food contact surfaces. This has been addressed in a variety of ways. Recently there has been interest in coupling ATP bioluminescence assays with immunomagnetic separation 28, 29 but the main disadvantage of this is the possibility of non-specific binding of microorganisms to the paramagnetic beads used in the technique. A commercial system has appeared that utilises host-specific bacteriophage to lyse target bacteria and the adenylate kinase that is released can be assayed using a bioluminescent method30. This forms the basis of the fastrAK™ assay supplied by Alaska Food Diagnostics.
Bacteriophages have been used in other ways to detect pathogens using a bioluminescent platform. Several researchers have used bacteriophage modified to carry luminescent or fluorescent reporter genes31. When the host cell is infected by the phage, the reporter genes are expressed and light is emitted. It has been shown that this method can be used for direct detection of bacteria on food surfaces32. As well as the structural genes encoding the enzymes involved in the bioluminescence reaction, researchers have constructed phage containing the luxI gene which encodes a signalling compound that then induces several other genes, luxCDABE, whose activation leads to bioluminescence in a reporter bacterium present on a biosensor33. It has also been proposed that biotinylated phage coupled with quantum-dot nanocomplexes can be used to detect bacteria34. We are currently investigating methods for the immobilisation of phage that will make it possible to concentrate and detect bacteria in a single step35.
Biosensors and other applications
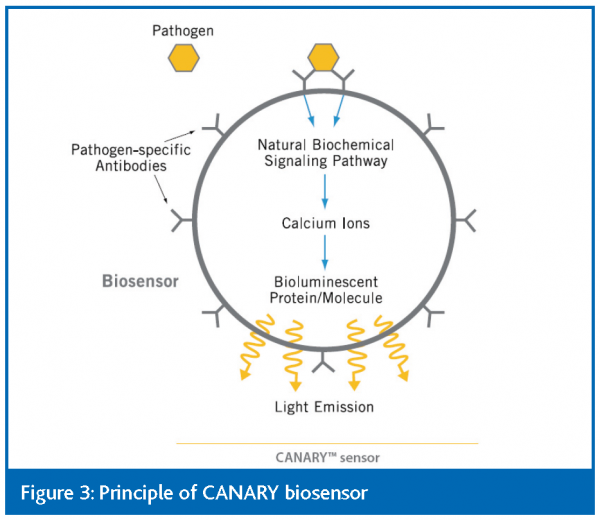
An interesting application of bioluminescence is the ‘CANARY’ biosensor. This involves a macrophage cell that exhibits pathogen-specific antibodies on its surface36. When the target cell binds to the antibody natural signalling pathways are triggered in the cell that can be detected by aequorin, which is expressed by the macrophage (Figure 3). The biosensor is being marketed by Innovative Biosensors, Inc. and has been applied to the detection of E. coli O157:H7 in ground beef.
Bioluminescent assays have also been developed for the detection of toxicants, antibiotics and cellular metabolites.
Much recent research has been focused on in vivo imaging of bacterial pathogens that have been genetically modified to carry luciferase genes. The efficacy of treatments to prevent foodborne infections can be determined using this technique37.
As well as building on current applications, there are several new and exciting opportunities offered to microbiologists by bioluminescence.




References
- Hastings,J.W. and Johnson,C.H. (2003) Bioluminescence and chemiluminescence. Biophotonics, Pt A 360, 75-104.
- Griffiths,M.W. (1996) The role of ATP bioluminescence in the food industry: New light on old problems. Food Technology 50, 62-72.
- Griffiths,M.W. (2000) How novel methods can help discover more information about foodborne pathogens. Canadian Journal of Infectious Diseases 11, 142-153.
- Nicolas,J.C. (1994) Applications of Low-Light Imaging to Life Sciences. Journal of Bioluminescence and Chemiluminescence 9, 139-144
- Bell,C., Bowles,C.D., Toszeghy,M.J.K. and Neaves,P. (1996) Development of a hygiene standard for raw milk based on the lumac ATP-bioluminescence method. International Dairy Journal 6, 709-713.
- Frundzhyan,V.G., Brovko,L.Y., Babunova,V.S., Kartashova,V.M. and Ugarova,N.N. (1999) A bioluminescence assay of total bacterial contamination of fresh milk. Applied Biochemistry and Microbiology 35, 321-327.
- Griffiths,M.W. (1993) Applications of Bioluminescence in the Dairy-Industry. Journal of Dairy Science 76, 3118-3125.
- Reybroeck,W. and Schram,E. (1995) Improved Filtration Method to Assess Bacteriological Quality of Raw-Milk Based on Bioluminescence of Adenosine-Triphosphate. Netherlands Milk and Dairy Journal 49, 1-14
- Samkutty,P.J., Gough,R.H., Adkinson,R.W. and McGrew,P. (2001) Rapid assessment of the bacteriological quality of raw milk using ATP bioluminescence. Journal of Food Protection 64, 208-212.
- Sutherland,A.D., Bell,C., Limond,A., Deakin,J. and Hunter,E.A. (1994) The Biotrace Method for Estimating Bacterial Numbers in Milk by Bioluminescence. Journal of the Society of Dairy Technology 47, 117-120.
- Bautista,D.A., Vaillancourt,J.P., Clarke,R.A., Renwick,S. and Griffiths,M.W. (1994) Adenosine-Triphosphate Bioluminescence As A Method to Determine Microbial Levels in Scald and Chill Tanks at A Poultry Abattoir. Poultry Science 73, 1673-1678.
- Bautista,D.A., Vaillancourt,J.P., Clarke,R.A., Renwick,S. and Griffiths,M.W. (1995) Rapid Assessment of the Microbiological Quality of Poultry Carcasses Using Atp Bioluminescence. Journal of Food Protection 58, 551-554
- Ellerbroek,L. and Lox,C. (2004) Investigation on the use of neck skin for microbial process control of fresh poultry meat with the ATP Bioluminescence method. Deutsche Tierarztliche Wochenschrift 111, 181-184.
- Siragusa,G.R., Dorsa,W.J., Cutter,C.N., Perino,L.J. and Koohmaraie,M. (1996) Use of a newly developed rapid microbial ATP bioluminescence assay to detect microbial contamination on poultry carcasses. Journal of Bioluminescence and Chemiluminescence 11, 297-301.
- Siragusa,G.R., Cutter,C.N., Dorsa,W.J. and Koohmaraie,M. (1995) Use of A Rapid Microbial Atp Bioluminescence Assay to Detect Contamination on Beef and Pork Carcasses. Journal of Food Protection 58, 770-775.
- Werlein,H.D. (2001) Comparison of destructively and rinsing gained samples to determine TVC of pig carcasses by bioluminescence. Meat Science 59, 165-168.
- Ukuku,D.O., Sapers,G.M. and Fett,W.F. (2005) ATP bioluminescence assay for estimation of microbial populations of fresh-cut melon. Journal of Food Protection 68, 2427-2432.
- Ukuku,D.O., Pilizota,V. and Sapers,G.M. (2001) Bioluminescence ATP assay for estimating total plate counts of surface microflora of whole cantaloupe and determining efficacy of washing treatments. Journal of Food Protection 64, 813-819.
- Hawronskyj,J.M. and Holah,J. (1997) ATP: A universal hygiene monitor. Trends in Food Science & Technology 8, 79-84.
- Moore,G. and Griffith,C. (2002) A comparison of traditional and recently developed methods for monitoring surface hygiene within the food industry: an industry trial. International Journal of Environmental Health Research 12, 317-329.
- Green,E.A., Russell,S.M. and Fletcher,D.L. (1998) Effect of chemical sanitizing agents on ATP bioluminescence measurements. Journal of Food Protection 61, 1013-1017.
- Green,T.A., Russell,S.M. and Fletcher,D.L. (1999) Effect of chemical cleaning agents and commercial sanitizers on ATP bioluminescence measurements. Journal of Food Protection 62, 86-90
- Velazquez,M. and Feirtag,J.M. (1997) Quenching and enhancement effects of ATP extractants, cleansers, and sanitizers on the detection of the ATP bioluminescence signal. Journal of Food Protection 60, 799-803.
- Hattori,N., Kajiyama,N., Maeda,M. and Murakami,S. (2002) Mutant luciferase enzymes from fireflies with increased resistance to benzalkonium chloride. Bioscience Biotechnology and Biochemistry 66, 2587-2593.
- Satoh,T., Kato,J., Takiguchi,N., Ohtake,H. and Kuroda,A. (2004) ATP amplification for ultrasensitive bioluminescence assay: Detection of a single bacterial cell. Bioscience Biotechnology and Biochemistry 68, 1216-1220.
- Squirrel,D.J., Price,R.L. and Murphy,M.J. (2002) Rapid and specific detection of bacteria using bioluminescence. Analytica Chimica Acta 457, 109-114.
- Sakakibara,T., Murakami,S. and Imai,K. (2003) Enumeration of bacterial cell numbers by amplified firefly bioluminescence without cultivation. Analytical Biochemistry 312, 48-56.
- Aytac,S.A., Mercanoglu,B. and Ozbas,Z.Y. (2001) Enumeration of Escherichia coli O157 : H7 by using immunomagnetic separation and ATP bioluminescence in buffer solution. Archiv fur Lebensmittelhygiene 52, 124-126.
- Sun,W., Khosravi,F., Albrechtsen,H., Brovko,L.Y. and Griffiths,M.W. (2002) Comparison of ATP and in vivo bioluminescence for assessing the efficiency of immunomagnetic sorbents for live Escherichia coli O157 : H7 cells. Journal of Applied Microbiology 92, 1021-1027.
- Blasco,R., Murphy,M.J., Sanders,M.F. and Squirrell,D.J. (1998) Specific assays for bacteria using phage mediated release of adenylate kinase. Journal of Applied Microbiology 84, 661-666.
- Goodridge,L. and Griffiths,M. (2002) Reporter bacteriophage assays as a means to detect foodborne pathogenic bacteria. Food Research International 35, 863-870.
- Chen,J. and Griffiths,M.W. (1996) Salmonella detection in eggs using Lux(+) bacteriophages. Journal of Food Protection 59, 908-914.
- Ripp,S., Jegier,P., Birmele,M., Johnson,C.M., Daumer,K.A., Garland,J.L. and Sayler,G.S. (2006) Linking bacteriophage infection to quorum sensing signalling and bioluminescent bioreporter monitoring for direct detection of bacterial agents. Journal of Applied Microbiology 100, 488-499.
- Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, Giulian G, Merril C, Nagashima K and Adhya S (2006) High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. U. S. A 103, 4841-4845.
- Sun,W., Brovko,L. and Griffiths,M. (2000) Use of bioluminescent Salmonella for assessing the efficiency of constructed phage-based biosorbent. Journal of Industrial Microbiology & Biotechnology 25, 273-275.
- Rider,T.H., Petrovick,M.S., Nargi,F.E., Harper,J.D., Schwoebel,E.D., Mathews,R.H., Blanchard,D.J., Bortolin,L.T., Young,A.M., Chen,J. and Hollis,M.A. (2003) A B Cell-Based Sensor for Rapid Identification of Pathogens. Science 301, 213-215.
- Brovko,L.Y., Vandenende,C., Chu,B., Ng,K.Y., Brooks,A. and Griffiths,M.W. (2003) In vivo assessment of effect of fermented milk diet on course of infection in mice with bioluminescent Salmonella. Journal of Food Protection 66, 2160-2163.









