Use of predictive microbiology in the food industry
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 16 November 2007 | Jeanne-Marie Membré, Unilever | No comments yet
The goal of predictive microbiology is to provide useful predictions about the microbial behaviour in food systems. Predictive microbiology combines “the disciplines of food microbiology, engineering and statistics” (Schaffner and Labuza, 1997).
The goal of predictive microbiology is to provide useful predictions about the microbial behaviour in food systems. Predictive microbiology combines “the disciplines of food microbiology, engineering and statistics” (Schaffner and Labuza, 1997).
The goal of predictive microbiology is to provide useful predictions about the microbial behaviour in food systems. Predictive microbiology combines “the disciplines of food microbiology, engineering and statistics” (Schaffner and Labuza, 1997).
Applications of predictive microbiology in an industrial context are wide but could be summarised into three groups of activities:
- Product innovation: Assessing speed of microbial proliferation, growth limits, or inactivation rate associated to particular food formulation/process conditions in order to develop new products and processes, reformulate existing products, and determine storage conditions and shelf-life.
- Operational support: Supporting food safety decisions that need to be made when implementing or running a food manufacturing operation, such as designing in-factory heating regimes, setting critical control points (CCPs) in HACCP, assessing impact of process deviations on microbiological safety and quality of food products.
- Incident support: Estimating the impact on consumer safety or product quality in case of problems with products on the market.
The latter is also a relevant application in governmental or public context, next to the application of predictive microbiology in Microbiological Risk Assessments (MRAs). In MRAs, the public health risk of hazards potentially associated with particular food products or food groups is quantified (Buchanan and Whiting, 1996;Walls and Scott, 1997; Coleman, 2003).
A quick historical tour
Predictive microbiology is an essential element of food microbiology (Baranyi, 2005). It is a relatively recent development considering that food preservation such as salting, drying and fermentation have been carried out for thousands of years.
Early examples of the application of scientific principles to food preservation include Pasteur’s work on the specificity of undesirable fermentations in wine and the supply of lactic starter cultures by Hansen in Denmark at the end of the 19th century. The Arrhenius equation (1889) and the Bigelow model (1921) were used to mathematically describe the effect of temperature on heat resistant microorganisms. The botulinum cook, developed some 90 years ago, may have been established on the basis of the first predictive model, albeit only recently recognised as such, and has found widespread utility in the food industry (McMeekin et al., 2002).
The 1980s saw a marked increase in interest in predictive microbiology as a result of some major food poisoning outbreaks caused by both ‘traditional’ pathogens and foods (e.g. Salmonella in eggs) and emerging pathogens with unusual characteristics (e.g. Listeria monocytogenes able to grow at refrigeration temperatures). The resulting public and political awareness contributed to the prioritisation of food safety research by governments in the USA, Europe, Australia and New Zealand. The concomitant maturation of information technology and step-change increase in computer technology strongly enabled the development of the mathematics and computer power necessary for predictive modelling.
In the last two decades, predictive microbiology has established itself as a scientific disciplinary in its own right, with regular international multi-disciplinary conferences, scientific articles in peer-reviewed journals, books and internet websites.
The ultimate goal of predictive microbiology is to bridge the knowledge gap between the food (e.g preservative factors) and the biology (e.g. molecular mechanism).
Models and model development
Today, a great variety of models exist to describe the behaviour of populations of microorganisms (e.g. growth, death, survival, interactions with other microbes) in fluid and structured foods. There are different classes of models, referred to as primary or secondary models.
Primary models describe the behaviour of a bacterial population that is exposed to a specific environmental condition (e.g. a specific temperature, pH, water activity, preservative concentration) as a function of time. They can be applied to describe or simulate the kinetics of, for instance, growth or death in the population of micro-organisms.
Secondary models take the information captured in primary models describing a set of related specific environmental conditions together. For instance, they express how the maximum rate of growth in a population changes with increasing temperature (Figure 1). Other models used in food industry capture the time required for a 10 fold log reduction in microorganisms or the length of the lag phase to growth in relation to changes, for factors such as pH, storage temperature, and concentration of salts or sugars, and preservatives.
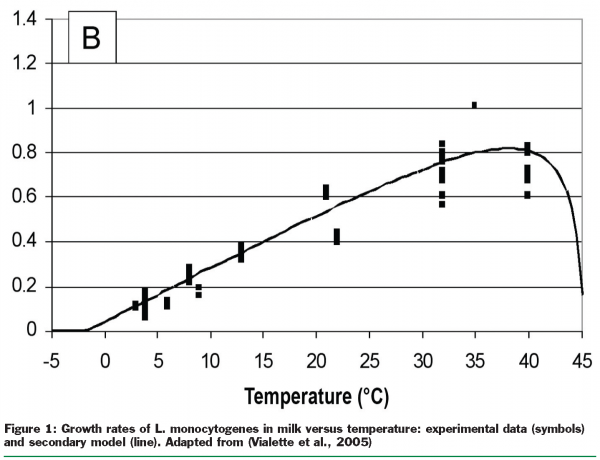

As some factors influencing the behaviour of microorganisms are not always unique when comparing similar food production and marketing systems at a certain point, models that are able to capture that variability may reflect reality better than models that more or less assume factors as single values. The so-called “probabilistic modelling approach” considers the impact of ranges of values on microorganisms and the probabilities associated with those impacts. Figure 2 illustrates the variation in temperature by showing the differences in temperature within a package of food that has been cooled down to refrigeration temperature.
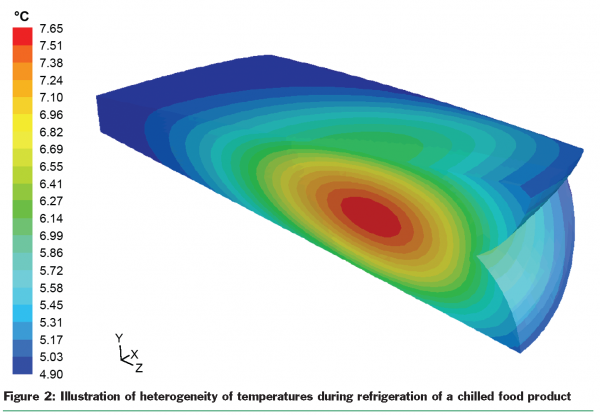

Next to the variability of model inputs at a given point in a system. Factors can significantly and intentionally vary over time, for instance, in the chilled food supply chain (Figure 2). In food industry, there are only very few situations where model inputs can be considered static, so have no variation with time. In most cases, the conditions influencing microorganisms are quite dynamic. Thermal processes such as pasteurisation or sterilisation are generally dynamic processes. Likewise, when products go through a food supply chain they are exposed to dynamic temperatures, sometimes being kept refrigerated and at other times transported at ambient temperatures.
Using mathematical models it is relatively quick and easy to run a lot of different food safety scenarios, i.e. to predict what may happen to a population of micro-organisms in foods with different storage temperatures or over different times of storage, without actually needing to run these scenarios. Obviously, it takes time and resources to collect the information needed to build a model in the first place and the model is likely to make useful predictions only when it is describing what happens in reality quite closely.
Generally, it is good practice to start by using a simple model that roughly describes reality and further refine or improve the model as required for the usefulness of the predictions or as allowed within time or resource constraints. For example, models are often developed using laboratory broth systems, even when they are to be used for structured, multi-component foods. The predictions of the first model are then compared to what happens under a number of conditions in a real food product. When the model is very far off the mark, it may not be very useful. When it is not all that bad, further work on the model may be worthwhile.
By now, mathematical models have been developed for several pathogenic bacteria as well as for some spoilage microorganisms. Many of the models and related information is available through the internet.
Predictive microbiology software
Software systems, either developed for commercial reasons or freely available on the internet, comprise a number of different models and provide access to these through user friendly interfaces.
- Growth Predictor (http://www.ifr.ac.uk/Safety/ GrowthPredictor/) provides a set of models for predicting the growth of a number of pathogenic bacteria, spoilage bacteria and yeasts as a function of key factors such as temperature, pH and water activity. No survival or inactivation models are included in the current version of Growth Predictor (version 1.0).
- Pathogen Modeling Program (http://www.arserrc.gov/mfs/ PMP7_start.htm) likewise provides models for predicting growth of (mainly pathogenic) bacteria but also includes several other model types. The current version of PMP (version 7.0) provides growth models, non-thermal survival models, thermal inactivation models, Gamma irradiation models, cooling/growth models, a time-to-toxigenesis model, and time-to-turbidity models.
While both these software packages have been developed separately, there is a joint activity in the form of a database that is useful for predictive modelling applications. This database, namely ComBase (http://wyndmoor.arserrc.gov/ combase/), contains a very large set of data brought together by researchers in academia, research institutions and food industry over several decades. This data provides a wealth of information on the behaviour of microorganisms under different conditions and is continuously extended. Such information can well be used to validate the outcome of new predictive models or as a source of modelling data, i.e. allowing the development of a model without having to generate new experimental data.
- Seafood Spoilage Predictor (http://www.dfu.min.dk/micro/sssp/ Home/Home.aspx) has been developed to predict shelf-life of a particular food group, namely seafood, at constant and fluctuating temperature storage conditions. The software can read data from different types of temperature loggers and in this way evaluate the effect of fluctuating temperatures on the shelf-life of seafood.
- The Sym’Previus software (http://www.symprevius.net/) contains both a simulation tool and a database. In the simulation tool, four bacterial species are available: Listeria monocytogenes, Salmonella, Bacillus cereus and Escherichia coli. For each species, many strains can be selected according to their origin (e.g. from dairy products, meat products, or sea food products). The current version includes growth models, thermal inactivation models and growth or no growth boundary models. The database contains a significant amount of data on the behaviour of microorganisms under different conditions and is used in association with the simulation tool.
Predictive microbiology at Unilever
In conclusion, at a maturation age of 30 years, predictive microbiology is still a rather new and dynamic research discipline. The very intensive work and development over that time has brought to bear a good number of concepts, tools and applications that are useful for both public and private organisations dealing with food safety and quality. Unilever has been involved with predictive microbiology from early on and our modellers and food microbiologists have looked at a variety of scientific and technical aspects ranging from heat-treatment inactivation (Kilsby et al., 2000) and microbiological risk assessment (Brown et al., 1998;Van Gerwen and Gorris, 2004) to new risk management concepts. (Gorris, 2005; Membré et al., 2007)
Regarding product innovation, Unilever has established microbiological expert systems that are accessed from around the world through our intranet to help speed up time to market in a responsible way (Figure 3).
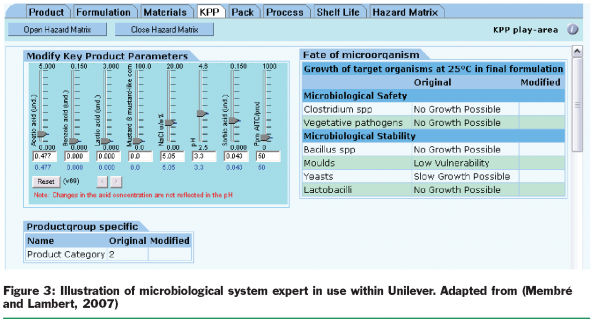

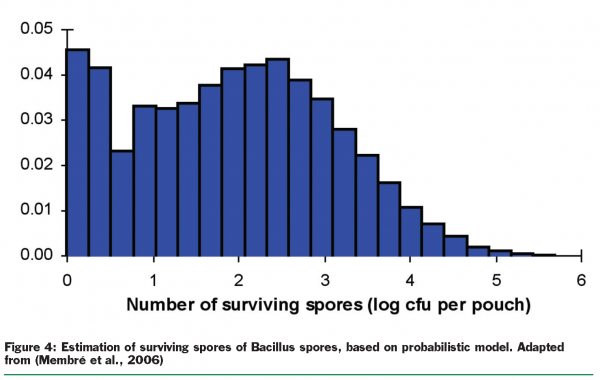
The expert system and specific tailor-made-models are used also for operational support. Particular models are used in probabilistic exposure assessment that allows increasingly realistic predictions (Figure 4). These applications benefit from the development of recent predictive models, such as those focused on individual lag phase or growth/no growth interfaces.

References
- Baranyi, J., 2005, Quantitative microbial ecology of food – Evolution of mathematical modelling in food microbiology: Acta Alimentaria, v. 34, no. 4, p. 335-337.
- Brown, M. H., K. W. Davies, C. M. P. Billon, C. Adair, and P. J. McClure, 1998, Quantitative microbiological risk assessment: principles applied to determining the comparative risk of salmonellosis from chicken products: Journal of Food Protection, v. 61, no. 11, p. 1446-1453.
- Buchanan, R. L., and R. C. Whiting, 1996, Risk assessment and predictive microbiology: Journal of Food Protection, p. 31-36.
- Coleman, M. E., 2003, Guest editorial: interactions of predictive microbiology and risk assessment: Risk Analysis, v. 23, p. 175-178.
- Gorris, L. G. M., 2005, Food safety objective: An integral part of food chain management: Food Control, v. 16, no. 9, p. 801-809.
- Kilsby, D. C., K. W. Davies, P. J. McClure, C. Adair, and W. A. Anderson, 2000, Bacterial thermal death kinetics based on probability distributions: The heat destruction of Clostridium botulinum and Salmonella Bedford: Journal of Food Protection, v. 63, no. 9, p. 1197-1203.
- McMeekin, T. A., J. Olley, D. A. Ratkowsky, and T. Ross, 2002, Predictive microbiology: towards the interface and beyond: International Journal of Food Microbiology, v. 73, no. 2-3, p. 395-407.
- Membré, J.-M., A. Amézquita, J. Bassett, P. Giavedoni, C. d. W. Blackburn, and L. G. M. Gorris, 2006, A probabilistic modeling approach in thermal inactivation: Estimation of postprocess Bacillus cereus spore prevalence and concentration: Journal of Food Protection, v. 69, no. 1, p. 118-129.
- Membré, J.-M., J. Bassett, and Gorris L.G.M., 2007, Applying the Food Safety Objective and related standards to thermal inactivation of Salmonella in poultry meat: Journal of Food Protection, v. 70, no. 9, p. 2036-2044.
- Membré,J-M, R J W Lambert. Application of predictive modelling techniques in industry: from food design up to risk assessment. 2007. Athens. 5th International Conference of Predictive Microbiology.
- Schaffner, D. W., and T. P. Labuza, 1997, Predictive microbiology: Where are we, and where are we going?: Food Technology, v. 51, no. 4, p. 95-99.
- Van Gerwen, S. J. C., and L. G. M. Gorris, 2004, Application of elements of microbiological risk assessment in the food industry via a tiered approach: Journal of Food Protection, v. 67, no. 9, p. 2033-2040.
- Vialette, M., A. Pinon, B. Leporq, C. Dervin, and J.-M. Membré, 2005, Meta-analysis of food safety information based on a combination of a relational database and a predictive modeling tool: Risk Analysis, v. 25, no. 1, p. 75-83.
- Walls, I., and V. N. Scott, 1997, Use of predictive microbiology in microbial food safety risk assessment: International Journal of Food Microbiology, v. 36, no. 2-3, p. 97-102.









