Molecularly imprinted polymers for selective extraction of mycotoxins in food
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 28 February 2008 | D. De Smet, S. De Saeger and C. Van Peteghem, Ghent University, Faculty of Pharmaceutical Sciences, Laboratory of Food Analysis, Belgium | No comments yet
This article highlights the possibility of using molecularly imprinted polymers (MIPs) for the selective extraction of mycotoxins in food, instead of immunoaffinity columns which are based on the binding of an analyte, to the corresponding antibody. An overview of MIPs, developed for mycotoxin analysis, will be presented.
This article highlights the possibility of using molecularly imprinted polymers (MIPs) for the selective extraction of mycotoxins in food, instead of immunoaffinity columns which are based on the binding of an analyte, to the corresponding antibody. An overview of MIPs, developed for mycotoxin analysis, will be presented.
This article highlights the possibility of using molecularly imprinted polymers (MIPs) for the selective extraction of mycotoxins in food, instead of immunoaffinity columns which are based on the binding of an analyte, to the corresponding antibody. An overview of MIPs, developed for mycotoxin analysis, will be presented.
Molecular imprinting
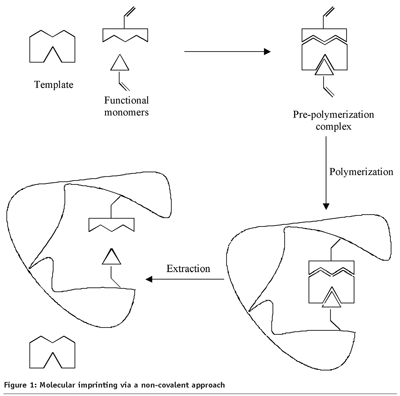
The technique of molecular imprinting is based on the formation of specific recognition sites in the synthesised polymer. These recognition sites mimic the binding sites of antibodies and enzymes, leading to MIPs with high selectivity and affinity to the target molecule. Besides good thermal and chemical stability, MIPs show many other advantages over antibodies, including low cost, ease of preparation, high mechanical strength, storage stability and repeated use, without loss of activity. Molecular imprinting is a process in which functional and cross-linking monomers are polymerised in the presence of the target molecule (template).
The template molecule is mixed with functional monomers, cross-linking monomers and initiator in a solvent. The template interacts with the functional monomer, forming a pre-polymerisation complex. Subsequently, the polymerisation will be initiated through UV light or heat. During polymerisation, the pre-polymerisation complex will be stabilised due to the copolymerisation of functional and cross-linking monomers, resulting in a highly cross-linked polymer. Finally, imprint binding sites, complementary in size and shape to the specific analyte, are formed by washing out the template molecule. In this way, the imprinted polymer exhibits a memory in the direction the target molecule or analogue structures.
MIPs can be mainly prepared by two imprinting techniques: non-covalent imprinting or covalent imprinting, depending on the interaction between template and monomer. In the covalent approach, the template will interact via reversible covalent bonding to the functional monomer. After the synthesis of the MIP, the imprint molecule will be removed by chemical cleavage, e.g. acid hydrolysis. A problem that arises, using this method, is that the covalent bonds with the analyte have to be reformed, resulting in slow association kinetics. During non-covalent imprinting, the template interacts with functional monomer, through non-covalent interactions, e.g. hydrogen bonds, ionic bonds, van der Waals forces and hydrophobic interactions. The formed pre-polymerisation complex is a reversible system at equilibrium with stability, depending on the affinity constants between the functional monomers and template, which causes heterogeneity in the imprinted binding sites. When applying the non-covalent method, an excess of functional monomer should be present to shift the equilibrium towards formation of the pre-polymerisation complex, which causes non-specific binding sites.
Of the two approaches, non-covalent imprinting (Figure 1) is mainly used because of a more simple synthesis of the pre-polymerisation complex, and because of a greater variety of functionality in the imprinted binding sites.
Remaining templates in the imprinted polymer, after washing, may result in template bleeding during solid-phase extraction, (SPE) giving erroneous results. To prevent template bleeding, a close structural analogue of the target molecule can be used during the imprinting process (Pichon et al., 2006) (Yan et al., 2006)8.
Application of MIPs in SPE (MIP-SPE)
MIPs have gained interest in many fields of chemistry and biology, especially in binding assays, sensors, chromatographic stationary phases and adsorbents for SPE. Today, commercial SPE sorbents, based on MIPs, are available for beta-agonists, riboflavin, chloramphenicol and triazines. MIPs used for MIP-SPE are commonly synthesised via bulk polymerisation, which gives a hard monolith that has to be crushed to produce particles in a desired size range (25-50 µm) (Pichon et al., 2006)8.
SPE is used for the extraction of compounds at trace levels from liquid samples or for the purification of extracts. Trace analysis of compounds from complex samples requires often very extensive and time consuming sample preparation, due to insufficient selectivity of traditional SPE sorbents. The principle of selective extraction on MIPs is similar to the extraction with immunoaffinity sorbents.
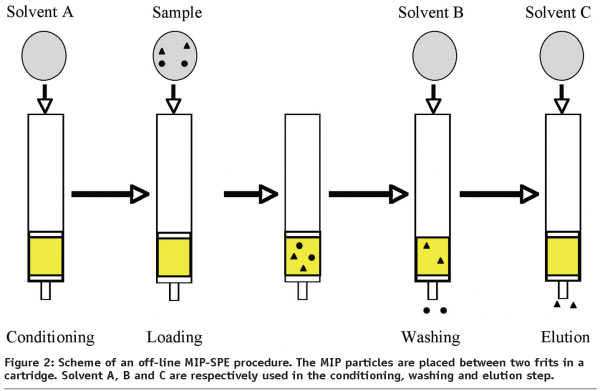
MIP-SPE procedures can be divided into off-line or on-line methods. In the off-line mode, a small amount of imprinted polymer (15-500 mg) is packed between two frits in cartridges. MIP-SPE takes place in four steps: conditioning, sample loading, washing and elution (Figure 2). During conditioning, the MIP cartridge is usually washed with eluting solvent to remove remaining analyte and then conditioned with loading solvent. The goal of this step is to minimise interferences and to activate the binding sites. Sample loading has to be optimised with the purpose of adsorbing quantitatively and selectively the analyte, by carefully choosing the loading solvent. The selection of loading solvent is dependent on the kind of interactions between analyte and monomers. If the binding of the target molecule is driven by electrostatic forces, e.g. ionic interactions and hydrogen bonding, a non-polar or low-polarity solvent should be selected, as loading solvent stabilises the interactions. After sample loading, the MIP cartridge is then washed to remove interfering compounds retained by non-specific interactions. To maximise the specific interactions between analyte and MIP, the washing step must be optimised in order to keep the target molecules retained on the MIP. At the end of the MIP-SPE procedure, the analyte has to be eluted. Desorption of the target molecules is achieved by percolating a solvent able to disrupt the specific interactions to recover them, quantitatively. To obtain high enrichment factors, it is necessary to use small volumes of solvent.
Furthermore, it is important to realise that MIPs are not intrinsically selective, but that the used MIP-SPE procedure confers its selectivity. From this point of view, the selection of solvents in the loading and washing steps should be carefully considered, as mentioned above. To control the selectivity of the MIP, a non-imprinted polymer (NIP) will be prepared, following the same synthesis procedure, as with the MIP, but without addition of template. By comparing the breakthrough percentages in the loading and washing steps of both MIP and NIP, one can have an idea of the specificity and selectivity.
To overcome the disadvantages (time-consuming procedure and higher possibility of analytical errors) of off-line MIP-SPE, an on-line MIP-SPE method can be carried out. This involves the automation of MIP-SPE, coupled to an analytical system and avoids sample manipulation between the preconcentration and analysis step. The use of an on-line procedure reduces the risk of contamination, the loss of analytes and improves the accuracy and the reproducibility. Other advantages over off-line MIP-SPE are smaller sample volumes and less time needed for pre-treatment of real samples.
In most cases, off-line MIP-SPE procedures are preferred because of the simple operation and the fact that more solvents and additives can be used without considering their influence on the subsequent chromatographic analysis (He et al., 2007) (Kaabi et al., 2007)3,4.
Mycotoxins
Mycotoxins are toxic secondary metabolites, produced by fungi growing under a wide range of climatic conditions, on agricultural commodities. Mycotoxins exhibit structural and chemical diversity, leading to different toxic effects: carcinogenic, neurotoxic, nephrotoxic, immunosuppressive and estrogenic effects. The toxic effect of mycotoxins depends on a number of factors: intake levels, duration of exposure and metabolism. Due to their various toxic effects, their occurrence in food, beverages and feed, good thermal and chemical stability, mycotoxins are a potential threat to human and animal health.
It is estimated that 25% of the world’s crop and 20% of crop production within the European Union may be contaminated with mycotoxins. Mycotoxins can be produced before, as well as after, the harvest. Due of the toxic effects taking place, several strategies have been developed to prevent the growth of mycotoxigenic fungi (Kabak et al., 2006) (Zöllner et al., 2006). Another issue is the presence of air-borne mycotoxins in mouldy buildings. After all, in any building that provides the proper mix of food, water and little air circulation mycotoxin producing fungi can be present (Assouline-Dayan et al., 2002)2,5,11.
The most important fungi producing mycotoxins belong to the genera Penicillium, Fusarium, Aspergillus and Alternaria. Aspergillus and Penicillium species produce mycotoxins during drying and storage, while Fusarium and Alternaria species contaminate food before or immediately after harvesting (Logrieco et al., 2003). Approximately 400 metabolites with toxic potential, produced by about 100 moulds, have been reported. Aflatoxins, trichothecenes, ochratoxin A, zearalenone, fumonisins, moniliformin and patulin are the most important, due to their frequent occurrence and their effects on human and animal health6.
Several countries have established legal regulations or recommendations for mycotoxins in food and feed. Under influence of maximum tolerable levels and guideline levels set by regulatory authorities, such as the European Commission and the US Food and Drug Administration, extensive efforts have been made to enable reliable analysis of mycotoxins in agricultural commodities and derived products (corn flakes, wine, apple juice…), down to the ppb level (Zöllner et al., 2006)11.
MIP-SPE of mycotoxins
Due to the complexity of food matrices, a clean-up step is necessary to analyse mycotoxins. Besides the conventional SPE-cartridges, e.g. SAX and C18, MIP-cartridges can be applied. MIPs prepared for the analysis of ochratoxin A (OTA), zearalenone (ZON) and moniliformin (MON) will be discussed briefly.
A two-dimensional SPE procedure on a C18 and a MIP cartridge was developed for the determination of the carcinogenic mycotoxin, OTA, found in red wine. However, similar performances were observed in control experiments in which the MIP was replaced by a NIP (Maier et al., 2004). To analyse OTA in wheat extracts, a MIP-SPE protocol was generated using a MIP based on N-phenylacrylamide. An inexpensive MIP-SPE-PE (pulsed elution) method was developed, without significant interference, from the matrix components, showing the applicability for rapid screening of OTA (Zhou et al., 2004)7,10.
A MIP prepared using 1-allylpiperazine as functional monomer was applied in a pressurised liquid extraction method to extract selectively ZON and α-zearalenol, a related metabolite, in cereal and swine feed. The combination of pressurised liquid extraction, MIP-SPE and liquid chromatography with fluorescence detection, offered good results for the determination of ZON and α-zearalenol: the obtained detection limits were lower than the EU maximum permitted levels (Urraca et al., 2006)9.
To extract MON in corn, a MIP synthesised with 2-(dimethylamino)ethyl methacrylate as functional monomer was applied as sorbent in a MIP-SPE procedure. Compared to NIPs, the imprinted polymers were capable to bind a larger amount of MON and were suitable for preconcentration of a MON spiked corn extract (Appell et al., 2007)1.
Conclusion
The general procedure, used to trace food contaminants, consists of the following steps: extraction, clean-up and a separating/detection/quantification step. With regard to the first steps, immunoaffinity columns exhibit excellent selectivity but this technique is rather expensive. Thus a new clean-up method with high specificity, which is cheap, environment-friendly and easy to prepare, is needed. Molecularly imprinted polymers, also called synthetic antibodies, can therefore be used in applications relying on specific molecular binding events. At the present time, MIPs are applied to carry out selective extractions or clean-up methods of analytes in complex matrices.
In spite of the advantages of MIP-SPE applied to food samples, only a few promising articles of MIPs developed to extract mycotoxins in complex matrices are published. Currently, available mycotoxin-selective MIPs are not yet competitive to immunosorbents and therefore more research is needed.


References
- Appell M., Kendra D.F., Kim E.K. and Maragos C.M. (2007). Synthesis and evaluation of molecularly imprinted polymers as sorbents of moniliformin. Food Additives and Contamininants, 24, 43-52.
- Assouline-Dayan Y., Leong A., Shoenfeld Y. and Gershwin M.E. (2002). Studies of sick building syndrome. IV. Mycotoxicosis. Journal of Asthma, 39, 191-201.
- He C., Long Y., Pan J., Li K. and Liu F. (2007). Application of molecularly imprinted polymers to solid-phase extraction of analytes from real samples. Journal of Biochemical and Biophysical Methods, 70, 133-150.
- Kaabi F.B. and Pichon V. (2007). Different approaches to synthesizing molecularly imprinted polymers for solid-phase extraction. LC GC Europe, 21, 407-413.
- Kabak B., Dobson A.D.W. and Var I. (2006). Strategies to prevent mycotoxin contamination of food and animal feed: a review. Critical Reviews in Food Science and Nutrition, 46, 593-619.
- Logrieco A., Bottalico A., Mulé G., Moretti A & Perrone G. (2003). Epidemiology of toxigenic fungi and their associated mycotoxins for some Mediterranean crops. European Journal of Plant Pathology, 109, 645-667.
- Maier N.M., Buttinger G., Welhartizki S., Gavioli E. and Lindner W. (2004). Molecularly imprinted polymer-assisted sample clean-up of ochratoxin A from red wine: merits and limitations. Journal of Chromatography B, 804, 103-111.
- Pichon V. and Haupt K. (2006). Affinity separations on molecularly imprinted polymers with special emphasis on solid-phase extraction. Journal of Liquid Chromatography & Related Technologies, 29, 989-1023.
- Urraca J.L., Marazuela M.D. and Moreno-Bondi M.C. (2006). Molecularly imprinted polymers applied to the clean-up of zearalenone and ·–zearalenol from cereal and swine feed sample extracts. Analytical and Bioanalytical Chemistry, 385, 1155-1161.
- Zhou S.N., Lai E.P.C. and Miller J.D. (2004). Analysis of wheat extracts for ochratoxin A by molecularly imprinted solid-phase extraction and pulsed elution. Analytical and Bioanalytical Chemistry, 378, 1903-1906.
- Zöllner P. and Mayer-Helm B. (2006). Trace mycotoxin analysis in complex biological and food matrices by liquid chromatography-atmospheric pressure ionisation mass spectrometry. Journal of Chromatography A, 1136, 123-169.









