Analysing barley to beer chain
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 23 May 2006 | Jari Rautio, Reetta Satokari, Kari Kataja,Anne Huuskonen, Heikki Vuokko,Arja Laitila,Annika Wilhelmson, Silja Home, Hans Söderlund and John Londesborough,VTT Technical Research Centre of Finland | No comments yet
VTT’s novel TRAC system is a rapid, accurate and economic method to quantitate specific messenger RNA molecules and other gene transcripts. In the barley-beer chain, it can be used to characterise yeast condition, to monitor fermentation and malting and to measure the levels of harmful and beneficial microorganisms through the whole process by analysing critical transcripts of yeast, barley and grain microflora.The knowledge can be used to control current processes and as an aid for designing new, improved processes.
Yeast must change during fermentations
The familiar and ancient fermentation of wort to beer is a complex process, in which yeast must adapt to sequential changes in carbon (glucose, maltose, maltotriose) and nitrogen sources; to depletion of essential nutrients (including oxygen) and to a variety of stresses, such as increasing levels of ethanol and sudden re-exposure to oxygen when yeast cropped from one fermentation is pitched into the next. Many changes in gene expression are expected as yeast struggles to adjust to its constantly changing environment. Quantitative knowledge about how transcription profiles change during the process offers the possibilities of modifying process conditions rationally in accord with yeast behaviour and routinely monitoring yeast condition in the factory.
VTT’s novel TRAC system is a rapid, accurate and economic method to quantitate specific messenger RNA molecules and other gene transcripts. In the barley-beer chain, it can be used to characterise yeast condition, to monitor fermentation and malting and to measure the levels of harmful and beneficial microorganisms through the whole process by analysing critical transcripts of yeast, barley and grain microflora.The knowledge can be used to control current processes and as an aid for designing new, improved processes. Yeast must change during fermentations The familiar and ancient fermentation of wort to beer is a complex process, in which yeast must adapt to sequential changes in carbon (glucose, maltose, maltotriose) and nitrogen sources; to depletion of essential nutrients (including oxygen) and to a variety of stresses, such as increasing levels of ethanol and sudden re-exposure to oxygen when yeast cropped from one fermentation is pitched into the next. Many changes in gene expression are expected as yeast struggles to adjust to its constantly changing environment. Quantitative knowledge about how transcription profiles change during the process offers the possibilities of modifying process conditions rationally in accord with yeast behaviour and routinely monitoring yeast condition in the factory.
VTT’s novel TRAC system is a rapid, accurate and economic method to quantitate specific messenger RNA molecules and other gene transcripts. In the barley-beer chain, it can be used to characterise yeast condition, to monitor fermentation and malting and to measure the levels of harmful and beneficial microorganisms through the whole process by analysing critical transcripts of yeast, barley and grain microflora.The knowledge can be used to control current processes and as an aid for designing new, improved processes.
Yeast must change during fermentations
The familiar and ancient fermentation of wort to beer is a complex process, in which yeast must adapt to sequential changes in carbon (glucose, maltose, maltotriose) and nitrogen sources; to depletion of essential nutrients (including oxygen) and to a variety of stresses, such as increasing levels of ethanol and sudden re-exposure to oxygen when yeast cropped from one fermentation is pitched into the next. Many changes in gene expression are expected as yeast struggles to adjust to its constantly changing environment. Quantitative knowledge about how transcription profiles change during the process offers the possibilities of modifying process conditions rationally in accord with yeast behaviour and routinely monitoring yeast condition in the factory.
Lager yeasts are hybrids formed by fusion of Saccharomyces cerevisiae and another yeast, possibly S. bayanus. For some years microarrays have been available for measuring transcripts of the complete set of just over 6000 S. cerevisiae genes. Microarrays have been used to study gene expression during laboratory and industrial scale wort fermentations. Usually, data collected at two or three time points through week long fermentations were compared1,2, though data collected daily have also been presented3. Here we describe a rapid, accurate and economical methodology, TRAC4 (TRanscriptional analysis with the aid of Affinity Capture), that facilitates frequent analysis of a chosen subset of genes. It can be used to follow transcription profiles in fermenting yeast or germinating barley and to quantitate specific harmful or beneficial microorganisms through the whole process, from the field microflora in grain to possible contaminants in bottled beer.
The TRAC methodology
For TRAC analysis of yeast, small samples of fermenting wort are rapidly filtered and the yeast transferred to liquid nitrogen. The cell membrane and enzymes that degrade mRNA are destroyed by thawing the yeast in the presence of a strong detergent. Transcript (mRNA) levels are measured from the resulting cell lysate without purification of RNA or conversion to cDNA. This avoids some sources of possible error. The next steps are conveniently performed by robot in a 96-well microtitre plate. Each chosen gene is recognised by a specific complementary probe, which is a fluorophore-labelled oligonucleotide.A pool of up to 20 probes with different lengths (usually 25-45 nucleotides) is added to each well. Hybridisation of probes and transcripts takes place in solution. Eukaryotic mRNA molecules have long polyA tails, so the probe-transcript complexes are captured via biotin-oligo-dT to magnetic beads coated with streptavidin, which tightly binds biotin. (To analyse nucleic acids without polyA tails, the target mixture is first biotinylated). Non-specifically bound probes and other material are washed away. The hydridisation, capture and washing are complete in 1-2 h. Specifically bound probes are then eluted and each pool is analysed by a 30 min capillary electrophoresis run, which resolves the probes mainly according to size. Separated probes are quantitated by their fluoresence.With one 96-well microtitre plate, a single yeast sample could be probed for 96 x 20 = 1920 distinct transcripts – approximately 30 per cent of the S. cerevisiae genome.Alternatively, the level of 20 chosen transcripts could be followed in 96 different yeast samples collected through fermentation. Because the material costs of analysing one 96-well plate are approximately 150 euros, and sufficient labelled probe for 10,000 analyses costs only around 50 euros, it is feasible to perform a large number of analyses.
Air causes fast changes in cropped yeast
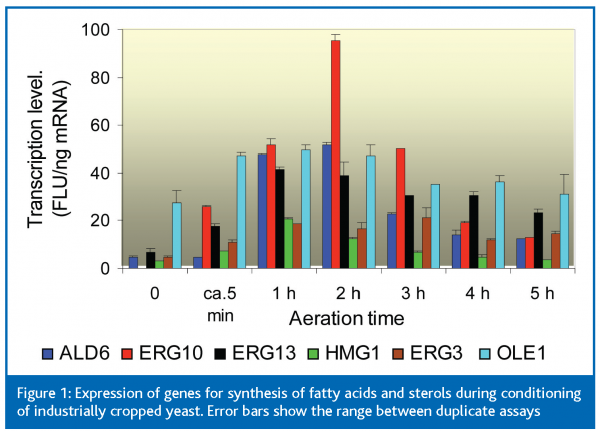
We used TRAC to help understand what happens when yeast cropped in a local brewery was first conditioned by aeration (to increase its contents of sterols and unsaturated fatty acids) and then pitched into very high gravity (VHG; 25 oP) wort on a laboratory scale. Figure 1 shows the changing expression, during conditioning, of several genes in the pathways of unsaturated fatty acid and sterol syntheses, from ALD6 (encoding the aldehyde dehydrogenase that forms cytosolic acetate) to OLE1 (for fatty acid desaturase) and ERG3 (for C5- sterol desaturase). Each of these genes, except for OLE1, were expressed at very low levels in the untreated cropped yeast (time point 0 in Fig 1). During the five minutes required to dilute the yeast slurry with two volumes of water and transfer it to the aeration vessel, the expression levels of all genes (except ALD6) increased by 2- to >20-fold (point ca. 5 min in Figure 1).After aeration for 1 h, expression of ALD6 had also increased >8-fold.After 2 h, transcript levels began to decline. These results show the great speed with which a slurry of cropped yeast responds when it is exposed to air.Although there is, of course, a lag between increases in mRNA and increases in proteins, the time courses of these changes help to understand how the anaerobically grown cropped yeast regenerates the sterols and unsaturated fats needed for vigorous fermentation.
Complex changes during early fermentation
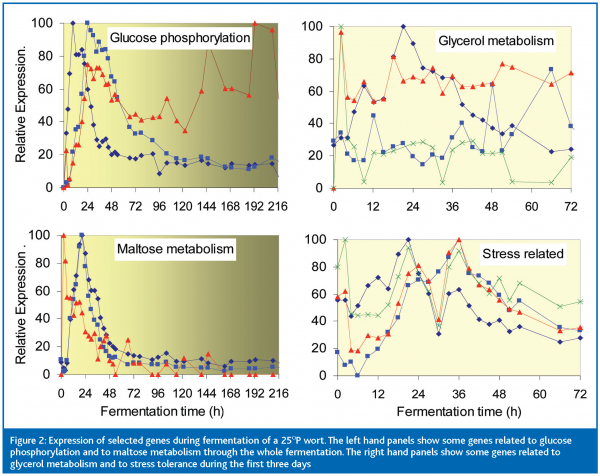
We then followed the expression of some 50 genes during fermentation of 25 oP wort. Figure 2 shows results for a selection of these genes. The top left panel shows that glucose phosphorylation genes were weakly expressed in the pitching yeast and then showed peaks of expression with different timings during early fermentation and the yeast growth phase (growth stopped at about 72 h). Only one of these genes was still strongly expressed at the end of fermentation. Genes involved in the transport and hydrolysis of maltose and maltotriose (bottom left panel) also showed well defined peaks and shoulders during early fermentation, while glucose was still present and the cells were still growing. However, by the time growth ceased, expression of each of these genes was again very low, so that metabolism of maltose and maltotriose during the rest of the fermentation and successfully complete attenuation of the wort depended upon events in early fermentation.
The yeast is very busy during the first 72 h. The right hand panels of Figure 2 show examples of this. Glycerol metabolism is important because intracellular glycerol increases to counter the high osmotic pressure of high gravity and VHG worts; glycerol is produced to maintain redox balance and glycerol contributes to the mouth feel of beer. Frequent TRAC assays show that the expression levels of the various genes involved change continually during the first 72 h. The same is true for expression of the group of stress-related genes shown in the bottom right panel of Figure 2. These results demonstrate that ‘snap shots’ of gene expression levels at single time points during the first two or three days of fermentation can give very misleading information about how the yeast is behaving. TRAC facilitates the frequent assays that reveal the dynamic changes in gene expression.
The TRAC technology can be applied to full scale industrial operations, requiring only that simple apparatus for rapid sample filtration is set up in the brewery. Probes can be designed that distinguish between the S. cerevisiae and corresponding S. bayanus genes of the hybrid lager yeasts. By focussing on a selection of important genes and collecting well replicated data, we expect to increase our knowledge of yeast behaviour through current brewery processes and in new process developments. This knowledge can guide improvements in yeast handling to increase efficiency and reproducibility.
Wide applications in the barley-beer chain
TRAC is applicable to virtually any complex mixture of nucleic acids and can be used in several ways through the barley to beer chain.As well as revealing yeast physiology in fermentation, it can reveal the physiology of germinating barley and of the field-derived microflora that also contribute, for better or worse, to the malting process. Sequence differences between 16S ribosomal RNAs of different bacteria mean that probes can be designed to recognise either total bacteria or particular groups of bacteria.VTT has used this approach to quantitate total bacteria and four phylogenetic groups of clostridia in a complex microbiological community5. The same approach can be developed to identify and quantitate harmful and beneficial microorganisms in grain and along the process chain through malting to bottling and distribution. For example, the level of potentially toxinproducing Fusarium spp in barley could be determined. Probes for transcripts of genes in the secondary metabolic pathways to toxins will not only indicate the presence of potentially dangerous microorganisms, but can tell whether they are actively producing toxins. Use of longer probes and PCR amplification of specifically bound probes increase specificity and make the method extremely sensitive6. The results can provide early warning of unwanted contaminants and their activities and be used to accept or reject material batches and to make decisions about process conditions.
VTT’s TRAC methodology is still a research tool, which can already be used to guide the operation and design of industrial processes.A further step now is to develop the technology into a convenient kit for routine monitoring of factory processes in real time.
Acknowledgements
The authors thank the Academy of Finland and the Finnish brewing and malting industry (PBL) for financial support.


References
- Higgins,V.J., Beckhouse,A.G.,Oliver,A.D., Rogers, P.J. and Dawes, I.W. (2003) Yeast genome-wide expression analysis identifies a strong ergosterol and oxidative stress response during the initial stages of an industrial lager fermentation.Appl. Environ.Microbiol. 69, 4777-4787.
- James, T.C., Campbell, S., Donnelly, D. and Bond, U. (2003) Transcription profile of brewery yeast under fermentation conditions. J.Appl.Microbiol. 94, 432-448.
- Olesen, K., Felding, T.,Gjermansen, C. and Hansen, J. (2002) The dynamics of the Saccharomyces carlsbergensis brewing yeast transcriptome during a production-scale lager beer fermentation. FEMS Yeast Research, 2, 561-573.
- Rautio, J.J., Kataja, K., Satokari, R., Penttilä,M., Söderlund, H. and Saloheimo,M. (2005) Rapid and multiplexed transcript analysis of microbial cultures using capillary electrophoresis-detectable oligonucleotide probe pools. J. Microbiol.Methods (Published online ahead of print, doi:10.1016/j.mimet.2005.08.010).
- Satokari, R.M., Kataja, K. and Söderlund, H. (2005) Multiplexed quantitation of bacterial 16S rRNA by solution hybridisation with oligonucleotide probes and affinity capture.Microbial Ecology, 50, 120-127.
- Kataja, K., Satokari, R.M.,Arvas,M., Takkinen, K. and Söderlund, H. (2006) A highly sensitive and multiplexed method for focused transcript analysis. J.Microbiol. Methods, in press.






