Detecting bacterial spores in soup manufacturing
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 10 September 2009 | A.C.M van Zuijlen & S.J.C.M Oomes, Unilever R&D; P. Vos, Check-Points B.V. and S. Brul, University of Amsterdam | No comments yet
Spores from mesophilic aerobic sporeforming bacteria (Bacillus) are sometimes able to survive the thermal process of commercial sterile products and sporadically cause spoilage or food poisoning. Because of an increasing demand for more fresh products, ideally the processing temperatures should be tailored to inactivate the actual spore load rather than applying worst case scenarios. In doing that, unnecessary loss of product quality can be prevented without running the risk that the product will spoil or cause safety issues.
Spores from mesophilic aerobic sporeforming bacteria (Bacillus) are sometimes able to survive the thermal process of commercial sterile products and sporadically cause spoilage or food poisoning. Because of an increasing demand for more fresh products, ideally the processing temperatures should be tailored to inactivate the actual spore load rather than applying worst case scenarios. In doing that, unnecessary loss of product quality can be prevented without running the risk that the product will spoil or cause safety issues.
Spores from mesophilic aerobic sporeforming bacteria (Bacillus) are sometimes able to survive the thermal process of commercial sterile products and sporadically cause spoilage or food poisoning. Because of an increasing demand for more fresh products, ideally the processing temperatures should be tailored to inactivate the actual spore load rather than applying worst case scenarios. In doing that, unnecessary loss of product quality can be prevented without running the risk that the product will spoil or cause safety issues.
In that respect, high heat resistant spores are of growing concern. These spores are introduced either into the product through ingredients with a high spore load or through growth and successive sporulation in the line during processing. To ensure an adequate level of thermal treatment to inactivate all spores in the process, their level in the ingredients must be known.
This paper describes new genomics based methods that allow for the rapid detection of bacterial spores in ingredients and semi final products.
Historically, the majority of soups and sauces have been produced directly for the consumer in small cans or glass jars. In large scale production of these products, the sterilisation process is based on inactivation of Clostridium botulinum (botulinum cook). However, in practice, for low acid soups and sauces (pH > 4.70) the processes are generally increased to F0 8 and more for inactivation of high heat resistant (Bacillus) sporeformers to obtain commercial sterile products. Spores are not totally eliminated but products are ambient stable for at least three years at temperatures of up to 37°C. In sporadic cases, however, spores were able to germinate and spoil the product. However, it is known from the literature that high heat resistant mesophilic spores, for example B. sporothermodurans1,2, are sometimes able to survive the sterilisation process. These spores are increasingly seen in processing environments3 and are capable of germination and outgrowth at ambient temperatures. Bacterial spores can enter the process through the ingredients, and may grow out during processing. The emerging vegetative cells are often able to sporulate again in-line during processing.
Spore heat resistance can be influenced by different factors such as incubation temperature, water content in the core4, and mineralisation3. Figure 1 explains the general process of soup production and indicates where spores are likely to enter the process.
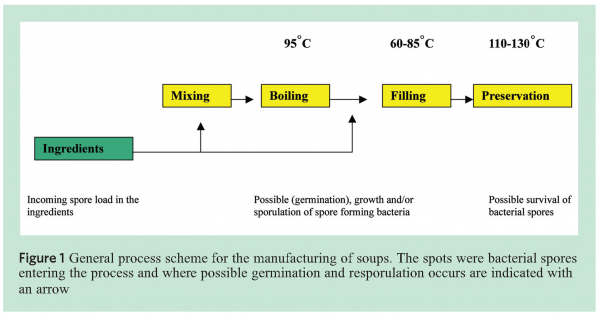

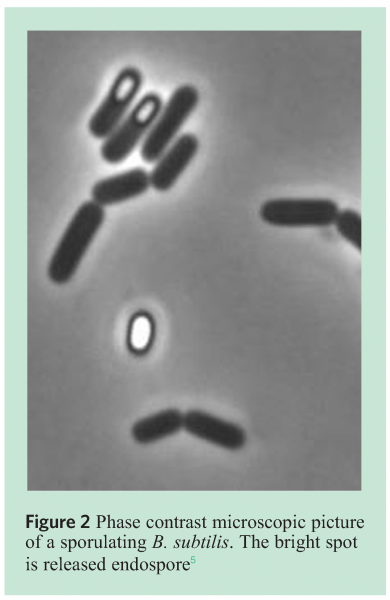
To be able to identify the various species of Bacilli, specific markers were developed and placed on a micro-array based platform. The markers covered, amongst others but not exclusively B. subtillis, B. sporthermodurans, B. licheniformis and B. cereus (see Figure 2[5]) for a typical example of a B. subtilis spore).

The output of this micro-array based platform gives information on the characteristics of the spore load which can be used to apply tailor-made thermal processing. This can lead to reduced processing and increased product quality.
Ingredients as source for bacterial spores
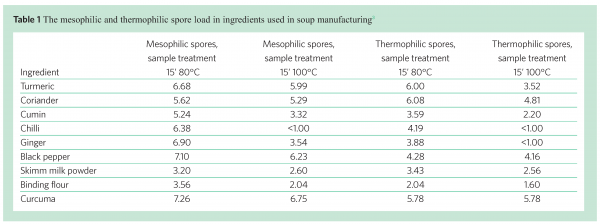
Some of the raw materials used in soup production, for instance spices and natural ingredients, are often heavily contaminated with bacterial spores. By performing spore counts after different heat treatments of the ingredient (80°C and 100°C), the number of high heat resistant spores can be measured. Examples of the spore load in different ingredients are given in Table 1. The composition of the ingredients may influence the heat resistance of the spores. Oomes et al.3 showed, for instance, that a high mineral, in particular calcium, content of ingredients used in soup manufacturing correlated with a high level of high heat resistant spores.

Growth of bacterial spores in a production line
In the production of canned soups and sauces, unknown numbers of Bacillus spores are generally introduced into the product through various ingredients. Production and cleaning regimes should control germination, growth and possibly re-sporulation during the allowed maximum production time to acceptable levels.
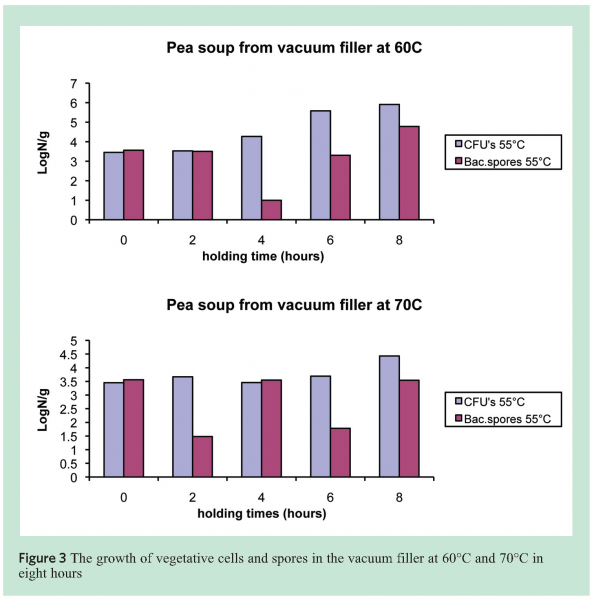
To establish maximum production and holding times in line, a number of soups were tested for microbial stability at 60, 70 and 80°C. These were assumed the critical temperatures at or above which the first adverse effects on outgrowth of aerobic and anaerobic spores may be observed. Analyses were performed with and without pasteurisation of the soup for 15 minutes at 80°C. The data in Figure 3 show the results for 60 and 70°C and it is obvious that sporeformers are present in the form of vegetative cells as well as spores after four to eight hours. Typing of the resulting strains showed that these were not exclusively thermophiles, but that sometimes also mesophilic sporeformers (B. licheniformis) could grow and sporulate.

Identifying relevant sporeforming bacteria and selection markers
To identify the species present in the ingredients and manufactured soups, different tools were used such as fatty acid analysis and 16S rRNA analysis. Fatty acid analysis is based on the lipids present in the cytoplasmic membranes of cells. The lipid composition is characteristic for individual microorganisms and can thus be used as a typing tool. Other molecular typing tools used were Ribotyping and Amplified Fragment Length Polymorphism (AFLP®). Ribotyping has been widely used for typing and characterisation of bacteria such as Salmonella6, Clostridium botulinum7 and Listeria monocytogenes8. Guillaume-Gentil9 reported on the use of ribotyping for the demonstration of genetic heterogeneity in B. sporothermodurans. RiboPrintR patterns are generated from restriction fragments of the ribosomal RNA genes. The DuPont Qualicon RiboPrinterTM is an established ribotyping instrument that is regularly used for the identification of bacteria. Sequence derived markers originating from the 23S or 16S genomic DNA may be used for more detailed strain analysis. Finally, the genome spanning AFLP® technology is often used to sub-type strains within a specific sporeformer species.
While reliable, all current tools to identify the spore load of ingredients are generally time consuming and indirect as they depend on isolation of the organisms of interest. What would be of major benefit to the industry is a tool that provides definitive results within eight hours. DNA-chip technology, coupled to proper nucleic acid isolation tools, could provide a proper platform for this.
Rapid testing technology
Polymerase Chain Reaction (PCR) based methods to characterise mesophilic Bacillus spores from UHT-milk have already been described by Hammer et al.10. It is thus possible to identify specific organisms, such as B. sporthermodurans, as reported by Scheldeman et al.11 and Montanari et al.12
Rapid screening for the detection of Bacillus spores in ingredients and in finished products was part of a Dutch national government subsidised project in the Ecology, Economy and Technology scheme. Relevant Bacilli were selected based on the similarity analyses performed with BioNumerics. Markers for commonly observed species (16S markers) and sometimes subspecies (genomic markers) in the case of B. subtilis were developed. Attempts were made to find specific markers for the group of heat resistant spores as well as specific markers for the genus Bacillus. The latter is necessary to identify strains which are not included yet in the set of markers for the selected Bacilli.
To perform these tests, DNA has to be extracted from a great variety of samples, varying from all kinds of ingredients to many different finished products. The extraction methods are based on centrifugation and on disintegration of the food matrix often using specific enzymes. After isolation of the spores from the different matrixes, the DNA has to be extracted. Extraction techniques were developed to obtain DNA with the highest possible yield from any given sample. An opportunity for the future would be to make use of a one step extraction such as described by Rossmanith et al.13

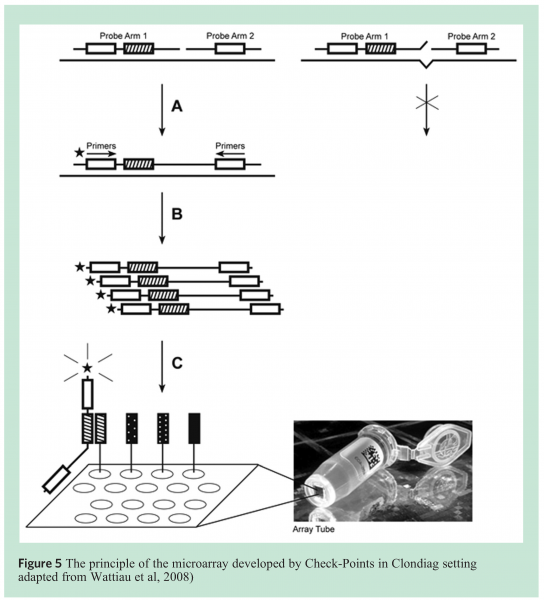
Check-Points has developed a generic microarray-based detection platform. This platform combines an efficient multiplex amplification method with detection on a diagnostic microarray (Array Tube technology, Clondiag, Germany). All Check-Points’ tests use the same set of reaction buffers for multiplex amplification and reagents and for microarray hybridisation and staining. Each assay consists of a set of specific DNA probes that generate a unique amplification product upon recognition of its target DNA sequence (Figure 5). Each probe also contains a unique ZIP oligonucleotide tag that is used for microarray detection of the corresponding amplification product to its complementary ZIP microarray tag. Amplification products hybridised to the microarray are visualised through colorimetric detection and images are recorded using an ArrayTube reader connected to a standard computer14 (Clondiag, Germany) (see Figure 5).

An assay for mesophilic and thermophilic Bacilli was developed allowing detection of many Bacillus species including Bacillus subtilis, sporothermodurans, coagulans, licheniformis, cereus and Geobacillus stearothermophilus. The assay was evaluated on a collection of approximately 100 strains of the above species and shown to detect and discriminate these species very well (unpublished results).
With a suitable rapid test system, samples can be screened for possible problematic Bacilli in raw materials (mainly dry ingredients and spices), and in products taken from the production line and after the thermal process.
The present status of these techniques does not allow this last step at this moment because surviving sporeformers will be well below the present detection level of 200 cells/spores per gram. This would require a detection level of one spore per packaging unit. The results from screening with a rapid test will enable application of the optimum thermal process without having to take into account the maximum calculated worst case spore load. This can result in reduced thermal processes and subsequently in better quality products, while still assuring ambient stability.
References
- Pettersson, B., Lembke, F., Hammer, P., Stackebrandt, E. and Priest, F.G. (1996), Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. International Journal of Systematic Bacteriology 46, 759-764.
- Scheldeman, P., Herman, L., Foster, L. S. and Heyndrickx, M., (2006), Bacillus sporothermodurans and other highly heat-resistant sporeformers in milk, Journal of Applied Microbiology 101, 542 – 555.
- Oomes, S.J.C.M., van Zuijlen, A.C.M., Hehenkamp, J.O., Witsenboer, H., van der Vossen, J.M.B.M. and Brul, S., (2007), The characterisation of Bacillus spores occurring in the manufacturing of (low acid) canned products. International Journal of Food Microbiology, 120, 85 – 94.
- Setlow, P and Johnson, E.A.(1997) Spores and their significants. In Food microbiology, fundamentals and frontiers. Editors Doyle and Beuchat, L.R. and Montville, T.J. pp30-62 ASM Press Washington, DC
- Ter Beek, A.S., 2009, Weak organic acid stress in Bacillus subtilis. Phd thesis. Chapter 1, 7-30
- Oscar, T.P. (1998), Identification and characterization of Salmonella isolates by automated Ribotyping, Journal of Food Protection, 61, 5, 519 – 524.
- Skinner, G. E., Gendel, S.M., Fingerhut, G. A., Solomon, H. A. and J. Ulaszek, J., (2000). Differentiation between types and strains of Clostridium botulinum by riboprinting. J. Food Prot. 63, 1347 – 1352.
- Gendel, S. M., and Ulaszek, J., (2000), Ribotype analysis of strain distribution in Listeria monocytogenes. J. Food Prot. 63:179-185.
- Guillaume-Gentil, O., Scheldeman, P., Marugg, J., Herman, L., Joosten, H., and Heyndrickx, M., (2002), Genetic heterogeneity in Bacillus sporothermodurans as demonstrated by ribotyping and repetitive extragenic palindromic PCR fingerprinting, Appl. Environ. Microbiol. 68, 4216 – 4224.
- Hammer, P., Lembke, F., Suhren, G. and Heeschen, W. (1995), Characterization of a heat resistant mesophilic Bacillus species affecting quality of UHT-milk – a preliminary report. Kieler Milchwirtschaftliche Forschungsberichte 47, 303-311.
- Scheldeman, P., Herman, L., Goris, J., De Vos, P. and Heyndrickx, M. (2002), Polymerase chain reaction identification of Bacillus sporothermodurans from dairy sources, Journal of Applied Microbiology, 92, 983 – 991
- Montanari, G., Borsari, A., Chiavari, C., Ferri, G., Zambonelli, C. and Grazia, L. (2004), Morphological and phenotypical characterization of Bacillus sporothermodurans. J. Appl. Microbiol. 97, 802-809.
- Rossmanith, P., Süβ, B., Wagner, W., Hein, I., (2007). Development of matrix lysis for concentration of gram positive bacteria from food and blood. Journal of Microbiological Methods, 69, 504-511
- Wattiau,P., Weijers, T., Andreoli, P., Schliker, C., Vander Veken, H., a, Maas, H.M.E., Verbruggen, A.J., Heck, M.E.O.C., Wannet, W.J., Imberechts, H., and Vos, P., (2008). Evaluation of the Premi®Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. International Journal of Food Microbiology 123 , 293-298









