PCR as a molecular method in the food industry
- Like
- Digg
- Del
- Tumblr
- VKontakte
- Buffer
- Love This
- Odnoklassniki
- Meneame
- Blogger
- Amazon
- Yahoo Mail
- Gmail
- AOL
- Newsvine
- HackerNews
- Evernote
- MySpace
- Mail.ru
- Viadeo
- Line
- Comments
- Yummly
- SMS
- Viber
- Telegram
- Subscribe
- Skype
- Facebook Messenger
- Kakao
- LiveJournal
- Yammer
- Edgar
- Fintel
- Mix
- Instapaper
- Copy Link
Posted: 22 February 2010 | Luca Cocolin, Paola Dolci & Kalliopi Rantsiou, DIVAPRA, Agricultural Microbiology and Food Technology Sector, Faculty of Agriculture, University of Turin | No comments yet
The presence of pathogens is a serious problem that industries producing foodstuffs have to face on a daily base. Foodborne pathogens can survive during processing or they can come in contact with the product due to recontamination or cross-contamination. Food products that contain pathogens represent a risk for human health since consumption of a contaminated product may lead to disease for the consumer. Consequently, there is a strong need for food industries to have methods for detection that are rapid, sensitive, specific and reliable.
The presence of pathogens is a serious problem that industries producing foodstuffs have to face on a daily base. Foodborne pathogens can survive during processing or they can come in contact with the product due to recontamination or cross-contamination. Food products that contain pathogens represent a risk for human health since consumption of a contaminated product may lead to disease for the consumer. Consequently, there is a strong need for food industries to have methods for detection that are rapid, sensitive, specific and reliable.
The presence of pathogens is a serious problem that industries producing foodstuffs have to face on a daily base. Foodborne pathogens can survive during processing or they can come in contact with the product due to recontamination or cross-contamination. Food products that contain pathogens represent a risk for human health since consumption of a contaminated product may lead to disease for the consumer. Consequently, there is a strong need for food industries to have methods for detection that are rapid, sensitive, specific and reliable.
The traditional microbiological methods do not fulfil the requirements requested by food business operators, primarily because of the time needed to obtain the result on the presence or absence of the specific pathogen. For Salmonella spp. there is a need for at least four to five days before confirming the presence of this bacterium in food products. Moreover, it is widely accepted and recognised that microbiological methods based on cultivation can present biases related with the use of laboratory media. Culture-dependent methods are not able to recover populations that entered the viable non culturable (VNC) state and cells that are numerically less abundant, which may be inhibited by the other major populations present in the system. The microbiological picture that is obtained by the use of methods based on cultivation may be completely different from the real ecological situation.
The application of PCR based methods in food microbiology allowed the development of protocols that are faster, more sensitive and more specific than traditional microbiological methods for the detection and identification of foodborne pathogens. Normally, the result on the presence or absence of a specific bacterium is obtained after three to four hours or at a maximum of 24 – 36 hours if an enrichment step is carried out. So far, different PCR protocols have been developed to detect specific foodborne pathogens. Some examples are: Listeria monocytogenes, Escherichia coli O157:H7, Salmonella spp., Campylobacter spp., Yersinia enterocoltica and Vibrio vulnificans. The target genes considered are virulence factors and the ribosomal RNA (rRNA gene). Commonly, PCR based methods consist of a selective enrichment prior to amplification, with the purpose of increasing the target cells, present at low number in the sample and of diluting food components that can inhibit the PCR reaction.
The PCR products obtained by amplification are routinely analysed by agarose gel electrophoresis. Gel electrophoresis is not always compatible with the number of samples to process for the quality control check. Moreover, it is characterised by poor sensitivity. Regardless, it remains a useful way of fast screening the samples. The problem of scarce sensitivity has been overcome with the use of more sophisticated protocols, based on the detection of the PCR product by using molecular probes. They are designed on specific target sequences and are hybridising to the amplified product, which is detected by colorimetric or chemioluminescence reactions on membranes or microtiter-plate wells.
The PCR protocols developed for the screening of clinical specimens at the beginning of the 1990’s began to be applied in the field of food microbiology. Researchers immediately expressed their interest in such techniques and this is demonstrated by the increasing number of scientific papers published in the last 10 years. In the period 1999-2009, an increase of 93.84 per cent in the number of papers published on the use of PCR for pathogen identification was recorded with respect to 1990-1998. This evidence should be explained by the increasing interest in these techniques by researchers all over the world and by the technological development that allowed different labs to implement these methods. The success of PCR in the field of food microbiology is undoubted. Moreover, the technological improvement allowed PCR to be applied not only as a qualitative technique but as a quantitative one as well. In this case, Real Time PCR is defined. With this method, the quantity of a specific DNA molecule, referable to a defined microorganism, can be determined.
The extensive development of molecular methods resulted in new tools that could be used for molecular characterisation of the pathogenic strains isolated from foodstuffs. Randomly amplified polymorphic DNA (RAPD)-PCR, repetitive bacterial DNA elements (Rep)-PCR and enterobacterial repetitive intergenic consensus (ERIC)-PCR are common practices in almost all the laboratories that deal with microbial diversity. At present, it is widely accepted that different methods for the identification and characterisation of foodborne pathogens, resulting in complementary information, are necessary. Molecular characterisation is applied to isolates that have been previously identified with other molecular techniques (i.e. specific PCR amplification, 16S rRNA gene sequencing) and intra-species differences are highlighted. Only strains belonging to the same species can be subjected to this kind of characterisation.
The methods that are more often used to characterise foodborne pathogens can be divided into methods that are exploiting PCR amplification and non-PCR methods. In the case of PCR-based methods, the differences in the strains to characterise can be detected either by exploiting primers that are annealing in various regions of the genome, thereby producing a band pattern that is representative for a strain, or by detecting differences in DNA sequences specifically amplified by PCR. RAPD-PCR and Rep-PCR are well-established techniques to look into molecular diversity. RAPD-PCR is characterised by an amplification process using a single random primer that will anneal in several portions of the genomic DNA and give amplification products when the forward and reverse annealing site are allowing extension by the DNA polymerase. The main drawback of this technique is its low reproducibility. The annealing step takes place at very low temperature (37-40°C) and for this reason, the same strain subjected to RAPD-PCR in different moments can produce different profiles. While it is possible to reach good intra-laboratory reproducibility if all the experimental steps are optimised and standardised, it is practically impossible to be able to compare results obtained in different laboratories. Concerning Rep-PCR, the primers used are specific to repetitive elements spread around the bacterial genomes and for this reason, the profiles obtained are highly specific for a species and they are highly reproducible as well. The techniques that allow strains characterisation based on differences in a DNA sequence of an amplified fragment are the restriction fragment length polymorphism (RFLP) analysis and the denaturing/temperature gradient gel electrophoresis (D/TGGE). With the RFLP approach, the differences in the DNA sequence are identified by using a restriction endonuclease, which is cutting the DNA in specific restriction sites, giving a specific pattern for a given strain. The D/TGGE methods detect differences in the sequences by analysing their denaturation behaviour. More specifically, as the DNA molecule encounters an appropriate denaturant concentration, a sequence- dependent partial denaturation of the double strand occurs. The change in the DNA conformation determines a reduction of the migration rate of the molecule, thereby DNA with different nucleotide sequences will show differential electrophoretic patterns. This approach was firstly used in environmental ecosystems and at the beginning of the 21st century, an impressive number of papers applying D/TGGE in the field of food microbiology and safety have been published.
Regarding the molecular methods that do not exploit an amplification step, the one most often used for pathogen characterisation is represented by the pulsed field gel electrophoresis (PFGE). Currently, this technique is considered the ‘gold standard’ for the characterisation of strains, since it is very precise, reproducible and reliable. In the field of foodborne pathogens, a database of PFGE profiles was created (PulseNet, http://www.cdc.gov/pulsenet/) thereby allowing epidemiologic analysis of foodborne disease outbreaks. This is possible because the results obtained by PFGE are comparable between different laboratories thanks to the reproducibility of the method. PFGE is an electrophoretic method that allows separation of the chromosomes for the yeasts and of macrorestriction fragments obtained from the genomic DNA for bacteria. The first step is the extraction of the DNA from the microorganism, which is embedded in agarose gel and subjected to lysis treatments in order to digest the cell wall. Once the DNA has been freed, it can be subjected directly to PFGE or it is digested with a restriction endonuclease that generally has the characteristic to be rare- cutting. In this way, a relatively small set of restriction fragments is obtained that are simply resolved in an agarose gel. The electrophoresis is carried out in specific chambers in which the electric field changes orientation. In this way, the DNA molecules are separated more efficiently than in conventional electrophoresis. The result obtained is a band pattern that is specific to the strain that was subjected to the PFGE analysis.
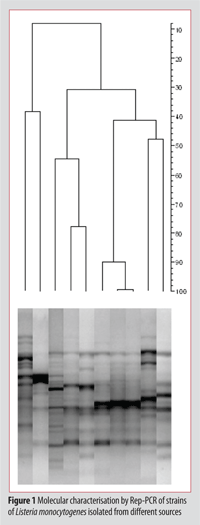
Molecular fingerprints obtained by profiling methods described above are usually processed by using image analysis software and dendrograms, as reported in Figure 1, are obtained. This process allows the differentiation of clusters containing strains of the same biotype.
In conclusion, nowadays we are experiencing important improvements in the food microbiology analysis, thanks to the availability of techniques that, as never before, can make significant information available regarding the safety of the foodstuffs. The invention of PCR, initially characterised by an extreme rapidity if compared with traditional microbiological methods, but not adequate for the enumeration of the cells of the pathogens, today has become a quantitative technique due to the possibility to monitor, in real time, the amplification of the PCR product.
Such DNA and RNA based assays offer versatility in reducing the incidence of false negative in the quality assurance screening measures, sensitivity in processing heterogeneous food matrices, specificity in differentiating among closely related bacterial strains, and speed, as in the case of many of the real time fluorescent chemistries currently on the market. Although some of these nucleic acid based technologies are unlikely to completely replace conventional culture, biochemical, or antibody-based testing methods, most offer significant potential for high throughput and reliability, advantages that offset the cost of initial equipment needs and ongoing maintenance or reagent expenses. Molecular diagnostics also offer a reliable initial screening tool so that follow-up, using conventional methods, is necessary only if a sample tests positive for the pathogen of interest. Lastly, the characterisation of foodborne pathogens can provide important information that can be used in order to understand their spread in terms of geographical distribution and source of isolation, thereby allowing a set up of intervention procedures to reduce or to eliminate specific types of pathogenic strains in the food chain.